A Beefy-R culture medium: Replacing albumin with rapeseed protein isolates
- PMID: 36965281
- PMCID: PMC10111969
- DOI: 10.1016/j.biomaterials.2023.122092
A Beefy-R culture medium: Replacing albumin with rapeseed protein isolates
Abstract
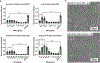
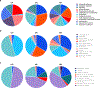
The development of cost-effective serum-free media is essential for the economic viability of cultured meat. A key challenge facing this goal is the high-cost of recombinant albumin which is necessary in many serum-free media formulations, including a recently developed serum-free medium for bovine satellite cell (BSC) culture termed Beefy-9. Here we alter Beefy-9 by replacing recombinant albumin with rapeseed protein isolate (RPI), a bulk-protein solution obtained from agricultural waste through alkali extraction (pH 12.5), isoelectric protein precipitation (pH 4.5), dissolution of physiologically soluble proteins (pH 7.2), and concentration of proteins through 3 kDa ultrafiltration. This new medium, termed Beefy-R, was then used to culture BSCs over four passages, during which cells grew with an average doubling time of 26.6 h, showing improved growth compared with Beefy-9. In Beefy-R, BSCs maintained cell phenotype and myogenicity. Together, these results offer an effective, low-cost, and sustainable alternative to albumin for serum-free culture of muscle stem cells, thereby addressing a key hurdle facing cultured meat production.
Keywords: Cellular agriculture; Cultured meat; Muscle satellite cells; Plant protein isolates; Serum-free media.
Copyright © 2023 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures




References
-
- Godfray HCJ et al. Meat consumption, health, and the environment. Science 361, eaam5324 (2018). - PubMed
-
- Mattick CS, Landis AE, Allenby BR & Genovese NJ Anticipatory Life Cycle Analysis of In Vitro Biomass Cultivation for Cultured Meat Production in the United States. Environ. Sci. Technol. 49, 11941–11949 (2015). - PubMed
-
- Post MJ et al. Scientific, sustainability and regulatory challenges of cultured meat. Nat. Food 1, 403–415 (2020).
-
- Tuomisto HL, Allan SJ & Ellis MJ Prospective life cycle assessment of a bioprocess design for cultured meat production in hollow fiber bioreactors. Sci. Total Environ. 851, 158051 (2022). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

