Genomic characterization of DICER1-associated neoplasms uncovers molecular classes
- PMID: 36966138
- PMCID: PMC10039902
- DOI: 10.1038/s41467-023-37092-w
Genomic characterization of DICER1-associated neoplasms uncovers molecular classes
Abstract
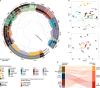
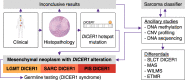
DICER1 syndrome is a tumor predisposition syndrome that is associated with up to 30 different neoplastic lesions, usually affecting children and adolescents. Here we identify a group of mesenchymal tumors which is highly associated with DICER1 syndrome, and molecularly distinct from other DICER1-associated tumors. This group of DICER1-associated mesenchymal tumors encompasses multiple well-established clinicopathological tumor entities and can be further divided into three clinically meaningful classes designated "low-grade mesenchymal tumor with DICER1 alteration" (LGMT DICER1), "sarcoma with DICER1 alteration" (SARC DICER1), and primary intracranial sarcoma with DICER1 alteration (PIS DICER1). Our study not only provides a combined approach to classify DICER1-associated neoplasms for improved clinical management but also suggests a role for global hypomethylation and other recurrent molecular events in sarcomatous differentiation in mesenchymal tumors with DICER1 alteration. Our results will facilitate future investigations into prognostication and therapeutic approaches for affected patients.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures