MicroRNAs emerging coordinate with placental mammals alter pathways in endometrial epithelia important for endometrial function
- PMID: 36968081
- PMCID: PMC10034127
- DOI: 10.1016/j.isci.2023.106339
MicroRNAs emerging coordinate with placental mammals alter pathways in endometrial epithelia important for endometrial function
Abstract
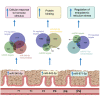
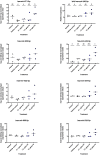
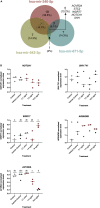
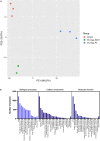
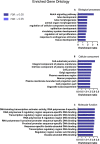
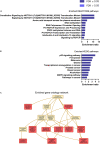
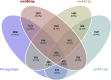
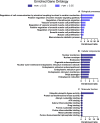
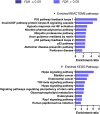
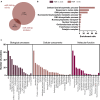
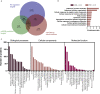
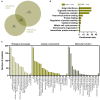
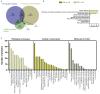
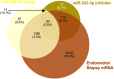
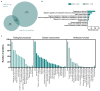
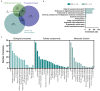
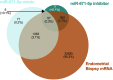
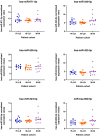
We tested the hypothesis that conserved placental mammal-specific microRNAs and their targets facilitate endometrial receptivity to implantation. Expression of miR-340-5p, -542-3p, and -671-5p was regulated by exposure of endometrial epithelial cells to progesterone (10 μg/ml) for 24 h coordinate with 1,713 of their predicted targets. Proteomic analysis of cells transfected with miRNA mimic/inhibitor (48 h: n = 3) revealed 1,745 proteins altered by miR-340-5p (mimic; 1,369, inhibitor; 376) of which 171 were predicted targets and P4-regulated. MiR-542-3p altered 2,353 (mimic; 1,378, inhibitor; 975) 100 which were mirDB predicted, including 46 P4-regulated. MiR-671-5p altered 1,744 proteins (mimic; 1,252, inhibitor; 492) 95 of which were predicted targets and 46 P4-regulated. All miRNAs were detected in luteal phase endometrial biopsies, irrespective of pregnancy outcomes. miR-340-5p expression increased in biopsies from individuals suffering previous and subsequent miscarriage compared to those with subsequent live birth. Dysfunction of these miRNAs and their targets contribute to endometrial-derived recurrent pregnancy loss.
Keywords: Biological sciences; Developmental biology; Embryology; Molecular biology.
© 2023 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





Similar articles
-
Progesterone suppresses podocalyxin partly by up-regulating miR-145 and miR-199 in human endometrial epithelial cells to enhance receptivity in in vitro models.Mol Hum Reprod. 2022 Oct 28;28(11):gaac034. doi: 10.1093/molehr/gaac034. Mol Hum Reprod. 2022. PMID: 36124965
-
Peritoneal fluid modifies the microRNA expression profile in endometrial and endometriotic cells from women with endometriosis.Hum Reprod. 2015 Oct;30(10):2292-302. doi: 10.1093/humrep/dev204. Epub 2015 Aug 25. Hum Reprod. 2015. PMID: 26307093
-
The miR-182-5p/NDRG1 Axis Controls Endometrial Receptivity through the NF-κB/ZEB1/E-Cadherin Pathway.Int J Mol Sci. 2022 Oct 14;23(20):12303. doi: 10.3390/ijms232012303. Int J Mol Sci. 2022. PMID: 36293154 Free PMC article.
-
[Differential expression of microRNA in eutopic endometrium tissue during implantation window for patients with endometriosis related infertility].Zhonghua Fu Chan Ke Za Zhi. 2016 Jun 25;51(6):436-41. doi: 10.3760/cma.j.issn.0529-567X.2016.06.007. Zhonghua Fu Chan Ke Za Zhi. 2016. PMID: 27356479 Chinese.
-
MicroRNAs in the Regulation of Endometrial Receptivity for Embryo Implantation.Int J Mol Sci. 2022 Jun 1;23(11):6210. doi: 10.3390/ijms23116210. Int J Mol Sci. 2022. PMID: 35682889 Free PMC article. Review.
Cited by
-
Research progress in the role of non-coding RNAs and embryo implantation.Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2023 Sept 28;48(9):1377-1387. doi: 10.11817/j.issn.1672-7347.2023.220485. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2023. PMID: 38044649 Free PMC article. Chinese, English.
-
Transcriptional response of endometrial cells to Insulin, cultured using microfluidics.Reprod Fertil. 2023 May 1;4(2):e210120. doi: 10.1530/RAF-21-0120. Online ahead of print. Reprod Fertil. 2023. PMID: 37200206 Free PMC article.
-
The role of microRNAs in pregnancies complicated by maternal diabetes.Clin Sci (Lond). 2024 Sep 18;138(18):1179-1207. doi: 10.1042/CS20230681. Clin Sci (Lond). 2024. PMID: 39289953 Free PMC article. Review.
-
Altered microRNA composition in the uterine lumen fluid in cattle (Bos taurus) pregnancies initiated by artificial insemination or transfer of an in vitro produced embryo.J Anim Sci Biotechnol. 2024 Sep 13;15(1):130. doi: 10.1186/s40104-024-01083-8. J Anim Sci Biotechnol. 2024. PMID: 39267128 Free PMC article.
-
Granulosa Cells-Related MicroRNAs in Ovarian Diseases: Mechanism, Facts and Perspectives.Reprod Sci. 2024 Dec;31(12):3635-3650. doi: 10.1007/s43032-024-01523-w. Epub 2024 Apr 9. Reprod Sci. 2024. PMID: 38594585 Review.
References
-
- Macklon N.S., Brosens J.J. The human endometrium as a sensor of embryo quality. Biol. Reprod. 2014;91:98. - PubMed
-
- Quenby S., Gallos I.D., Dhillon-Smith R.K., Podesek M., Stephenson M.D., Fisher J., Brosens J.J., Brewin J., Ramhorst R., Lucas E.S., et al. Miscarriage matters: the epidemiological, physical, psychological, and economic costs of early pregnancy loss. Lancet. 2021;397:1658–1667. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

