Complex Patterns of Genomic Heterogeneity Identified in 42 Tumor Samples and ctDNA of a Pulmonary Atypical Carcinoid Patient
- PMID: 36968225
- PMCID: PMC10035512
- DOI: 10.1158/2767-9764.CRC-22-0101
Complex Patterns of Genomic Heterogeneity Identified in 42 Tumor Samples and ctDNA of a Pulmonary Atypical Carcinoid Patient
Abstract
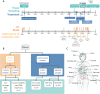
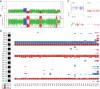
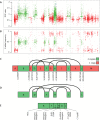
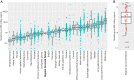
Tumor evolution underlies many challenges facing precision oncology, and improving our understanding has the potential to improve clinical care. This study represents a rare opportunity to study tumor heterogeneity and evolution in a patient with an understudied cancer type. A patient with pulmonary atypical carcinoid, a neuroendocrine tumor, metastatic to 90 sites, requested and consented to donate tissues for research. 42 tumor samples collected at rapid autopsy from 14 anatomically distinct sites were analyzed through DNA whole-exome sequencing and RNA sequencing, and five analyzed through linked-read sequencing. Targeted DNA sequencing was completed on two clinical tissue biopsies and one blood plasma sample. Chromosomal alterations and gene variants accumulated over time, and specific chromosomal alterations preceded the single predicted gene driver variant (ARID1A). At the time of autopsy, all sites shared the gain of one copy of Chr 5, loss of one copy of Chr 6 and 21, chromothripsis of one copy of Chr 11, and 39 small variants. Two tumor clones (carrying additional variants) were detected at metastatic sites, and occasionally in different regions of the same organ (e.g., within the pancreas). Circulating tumor DNA (ctDNA) sequencing detected shared tumor variants in the blood plasma and captured marked genomic heterogeneity, including all metastatic clones but few private tumor variants. This study describes genomic tumor evolution and dissemination of a pulmonary atypical carcinoid donated by a single generous patient. It highlights the critical role of chromosomal alterations in tumor initiation and explores the potential of ctDNA analysis to represent genomically heterogeneous disease.
Significance: DNA sequencing data from tumor samples and blood plasma from a single patient highlighted the critical early role of chromosomal alterations in atypical carcinoid tumor development. Common tumor variants were readily detected in the blood plasma, unlike emerging tumor variants, which has implications for using ctDNA to capture cancer evolution.
© 2023 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
No disclosures were reported.
Figures



References
-
- De Dosso S, Bajetta E, Procopio G, Cortinovis D, Buzzoni R, Catena L, et al. . Pulmonary carcinoid tumours: indolent but not benign. Oncology 2007;73:162–8. - PubMed
-
- Parker K, Coats E, Woodhouse B, Knowlton N, Schiedel O, Carroll R, et al. . The NETwork! registry: preliminary data suggests increasing incidence in a national study of neuroendocrine cancer in New Zealand (NZ). New Zealand Society Oncology; 2017; Auckland, New Zealand.
-
- Kahu TAT. He Pūrongo Mate Pukupuku o Aotearoa 2020, The State of Cancer in New Zealand 2020. Wellington: Te Aho o Te Kahu, Cancer Control Agency; 2021.
-
- Steuer CE, Behera M, Kim S, Chen Z, Saba NF, Pillai RN, et al. . Atypical carcinoid tumor of the lung: a surveillance, epidemiology, and end results database analysis. J Thorac Oncol 2015;10:479–85. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

