Misaligned Chromosomes are a Major Source of Chromosomal Instability in Breast Cancer
- PMID: 36968230
- PMCID: PMC10035514
- DOI: 10.1158/2767-9764.CRC-22-0302
Misaligned Chromosomes are a Major Source of Chromosomal Instability in Breast Cancer
Abstract
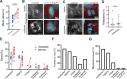
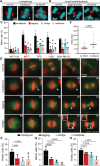
Chromosomal instability (CIN), the persistent reshuffling of chromosomes during mitosis, is a hallmark of human cancers that contributes to tumor heterogeneity and has been implicated in driving metastasis and altering responses to therapy. Though multiple mechanisms can produce CIN, lagging chromosomes generated from abnormal merotelic attachments are the major cause of CIN in a variety of cell lines, and are expected to predominate in cancer. Here, we quantify CIN in breast cancer using a tumor microarray, matched primary and metastatic samples, and patient-derived organoids from primary breast cancer. Surprisingly, misaligned chromosomes are more common than lagging chromosomes and represent a major source of CIN in primary and metastatic tumors. This feature of breast cancers is conserved in a majority of breast cancer cell lines. Importantly, though a portion of misaligned chromosomes align before anaphase onset, the fraction that remain represents the largest source of CIN in these cells. Metastatic breast cancers exhibit higher rates of CIN than matched primary cancers, primarily due to increases in misaligned chromosomes. Whether CIN causes immune activation or evasion is controversial. We find that misaligned chromosomes result in immune-activating micronuclei substantially less frequently than lagging and bridge chromosomes and that breast cancers with greater frequencies of lagging chromosomes and chromosome bridges recruit more stromal tumor-infiltrating lymphocytes. These data indicate misaligned chromosomes represent a major mechanism of CIN in breast cancer and provide support for differential immunostimulatory effects of specific types of CIN.
Significance: We surveyed the single-cell landscape of mitotic defects that generate CIN in primary and metastatic breast cancer and relevant models. Misaligned chromosomes predominate, and are less immunostimulatory than other chromosome segregation errors.
© 2023 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
J.B. Tucker reports grants from NIH during the conduct of the study. S.C. Bonema reports grants from NIH during the conduct of the study. M.E. Burkard reports grants from NCI and National Institute for General Medical Sciences during the conduct of the study; personal fees from Springer Nature and Novartis; grants and personal fees from Strata Oncology; grants from Abbvie, Arcus, Apollomics, Elevation Oncology, Endeavor, Genentech, Puma, Loxo Oncology/Lilly, Seagen outside the submitted work; in addition, M.E. Burkard has a patent to 14/727399 issued, a patent to US20200157531 issued, and a patent to P200189US01 issued. B.A. Weaver reports grants from NIH and DOD during the conduct of the study. No disclosures were reported by the other authors.
Figures



References
-
- Vasudevan A, Schukken KM, Sausville EL, Girish V, Adebambo OA, Sheltzer JM. Aneuploidy as a promoter and suppressor of malignant growth. Nat Rev Cancer 2021;21:89–103. - PubMed
-
- Laughney AM, Elizalde S, Genovese G, Bakhoum SF. Dynamics of tumor heterogeneity derived from clonal karyotypic evolution. Cell Rep 2015;12:809–20. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

