Genome-wide analysis of bromodomain gene family in Arabidopsis and rice
- PMID: 36968369
- PMCID: PMC10030601
- DOI: 10.3389/fpls.2023.1120012
Genome-wide analysis of bromodomain gene family in Arabidopsis and rice
Abstract
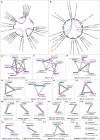
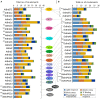
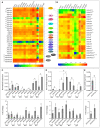
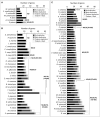
The bromodomain-containing proteins (BRD-proteins) belongs to family of 'epigenetic mark readers', integral to epigenetic regulation. The BRD-members contain a conserved 'bromodomain' (BRD/BRD-fold: interacts with acetylated-lysine in histones), and several additional domains, making them structurally/functionally diverse. Like animals, plants also contain multiple Brd-homologs, however the extent of their diversity and impact of molecular events (genomic duplications, alternative splicing, AS) therein, is relatively less explored. The present genome-wide analysis of Brd-gene families of Arabidopsis thaliana and Oryza sativa showed extensive diversity in structure of genes/proteins, regulatory elements, expression pattern, domains/motifs, and the bromodomain (w.r.t. length, sequence, location) among the Brd-members. Orthology analysis identified thirteen ortholog groups (OGs), three paralog groups (PGs) and four singleton members (STs). While more than 40% Brd-genes were affected by genomic duplication events in both plants, AS-events affected 60% A. thaliana and 41% O. sativa genes. These molecular events affected various regions (promoters, untranslated regions, exons) of different Brd-members with potential impact on expression and/or structure-function characteristics. RNA-Seq data analysis indicated differences in tissue-specificity and stress response of Brd-members. Analysis by RT-qPCR revealed differential abundance and salt stress response of duplicate A. thaliana and O. sativa Brd-genes. Further analysis of AtBrd gene, AtBrdPG1b showed salinity-induced modulation of splicing pattern. Bromodomain (BRD)-region based phylogenetic analysis placed the A. thaliana and O. sativa homologs into clusters/sub-clusters, mostly consistent with ortholog/paralog groups. The bromodomain-region displayed several conserved signatures in key BRD-fold elements (α-helices, loops), along with variations (1-20 sites) and indels among the BRD-duplicates. Homology modeling and superposition identified structural variations in BRD-folds of divergent and duplicate BRD-members, which might affect their interaction with the chromatin histones, and associated functions. The study also showed contribution of various duplication events in Brd-gene family expansion among diverse plants, including several monocot and dicot plant species.
Keywords: Arabidopsis thaliana; Oryza sativa; alternative splicing; bromodomain; bromodomain-containing genes; homology modeling; salt-stress response; tandem and block duplication.
Copyright © 2023 Abiraami, Sanyal, Misra and Saini.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures







References
-
- Airoldi C. A., Rovere F. D., Falasca G., Marino G., Kooiker M., Altamura M. M., et al. . (2010). The arabidopsis BET bromodomain factor GTE4 is involved in maintenance of the mitotic cell cycle during plant development. Plant Physiol. 152 (3), 1320–1334. doi: 10.1104/pp.109.150631 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

