An inflammation-related gene landscape predicts prognosis and response to immunotherapy in virus-associated hepatocellular carcinoma
- PMID: 36969014
- PMCID: PMC10033597
- DOI: 10.3389/fonc.2023.1118152
An inflammation-related gene landscape predicts prognosis and response to immunotherapy in virus-associated hepatocellular carcinoma
Abstract
Background: Due to the viral infection, chronic inflammation significantly increases the likelihood of hepatocellular carcinoma (HCC) development. Nevertheless, an inflammation-based signature aimed to predict the prognosis and therapeutic effect in virus-related HCC has rarely been established.
Method: Based on the integrated analysis, inflammation-associated genes (IRGs) were systematically assessed. We comprehensively investigated the correlation between inflammation and transcriptional profiles, prognosis, and immune cell infiltration. Then, an inflammation-related risk model (IRM) to predict the overall survival (OS) and response to treatment for virus-related HCC patients was constructed and verified. Also, the potential association between IRGs and tumor microenvironment (TME) was investigated. Ultimately, hub genes were validated in plasma samples and cell lines via qRT-PCR. After transfection with shCCL20 combined with overSLC7A2, morphological change of SMMC7721 and huh7 cells was observed. Tumorigenicity model in nude mouse was established.
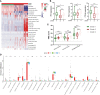
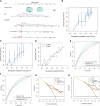
Results: An inflammatory response-related gene signature model, containing MEP1A, CCL20, ADORA2B, TNFSF9, ICAM4, and SLC7A2, was constructed by conjoint analysis of least absolute shrinkage and selection operator (LASSO) Cox regression and gaussian finite mixture model (GMM). Besides, survival analysis attested that higher IRG scores were positively relevant to worse survival outcomes in virus-related HCC patients, which was testified by external validation cohorts (the ICGC cohort and GSE84337 dataset). Univariate and multivariate Cox regression analyses commonly proved that the IRG was an independent prognostic factor for virus-related HCC patients. Thus, a nomogram with clinical factors and IRG was also constructed to superiorly predict the prognosis of patients. Featured with microsatellite instability-high, mutation burden, and immune activation, lower IRG score verified a superior OS for sufferers. Additionally, IRG score was remarkedly correlated with the cancer stem cell index and drug susceptibility. The measurement of plasma samples further validated that CCL20 upexpression and SLC7A2 downexpression were positively related with virus-related HCC patients, which was in accord with the results in cell lines. Furthermore, CCL20 knockdown combined with SLC7A2 overexpression availably weakened the tumor growth in vivo.
Conclusions: Collectively, IRG score, serving as a potential candidate, accurately and stably predicted the prognosis and response to immunotherapy in virus-related HCC patients, which could guide individualized treatment decision-making for the sufferers.
Keywords: drug sensitivity; hepatocellular carcinoma; immune; inflammation; tumor microenvironment; virus.
Copyright © 2023 Gao, Li and Huang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures








References
LinkOut - more resources
Full Text Sources
Research Materials

