Transcriptional control of ILC identity
- PMID: 36969171
- PMCID: PMC10033543
- DOI: 10.3389/fimmu.2023.1146077
Transcriptional control of ILC identity
Abstract
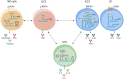
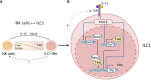
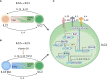
Innate lymphoid cells (ILCs) are heterogeneous innate immune cells which participate in host defense, mucosal repair and immunopathology by producing effector cytokines similarly to their adaptive immune cell counterparts. The development of ILC1, 2, and 3 subsets is controlled by core transcription factors: T-bet, GATA3, and RORγt, respectively. ILCs can undergo plasticity and transdifferentiate to other ILC subsets in response to invading pathogens and changes in local tissue environment. Accumulating evidence suggests that the plasticity and the maintenance of ILC identity is controlled by a balance between these and additional transcription factors such as STATs, Batf, Ikaros, Runx3, c-Maf, Bcl11b, and Zbtb46, activated in response to lineage-guiding cytokines. However, how interplay between these transcription factors leads to ILC plasticity and the maintenance of ILC identity remains hypothetical. In this review, we discuss recent advances in understanding transcriptional regulation of ILCs in homeostatic and inflammatory conditions.
Keywords: ILC identity; ILC plasticity; innate lymphoid cells; transcription factor; transcriptional regulation.
Copyright © 2023 Korchagina, Shein, Koroleva and Tumanov.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

