Maximum likelihood-based estimation of diffusion coefficient is quick and reliable method for analyzing estradiol actions on surface receptor movements
- PMID: 36970656
- PMCID: PMC10031098
- DOI: 10.3389/fninf.2023.1005936
Maximum likelihood-based estimation of diffusion coefficient is quick and reliable method for analyzing estradiol actions on surface receptor movements
Abstract
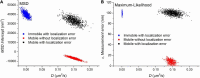
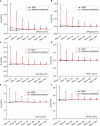
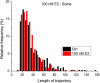
The rapid effects of estradiol on membrane receptors are in the focus of the estradiol research field, however, the molecular mechanisms of these non-classical estradiol actions are poorly understood. Since the lateral diffusion of membrane receptors is an important indicator of their function, a deeper understanding of the underlying mechanisms of non-classical estradiol actions can be achieved by investigating receptor dynamics. Diffusion coefficient is a crucial and widely used parameter to characterize the movement of receptors in the cell membrane. The aim of this study was to investigate the differences between maximum likelihood-based estimation (MLE) and mean square displacement (MSD) based calculation of diffusion coefficients. In this work we applied both MSD and MLE to calculate diffusion coefficients. Single particle trajectories were extracted from simulation as well as from α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor tracking in live estradiol-treated differentiated PC12 (dPC12) cells. The comparison of the obtained diffusion coefficients revealed the superiority of MLE over the generally used MSD analysis. Our results suggest the use of the MLE of diffusion coefficients because as it has a better performance, especially for large localization errors or slow receptor movements.
Keywords: MLE; diffusion coefficient; maximum likelihood; mean square displacement; receptor movements.
Copyright © 2023 Makkai, Abraham, Barabas, Godo, Ernszt, Kovacs, Kovacs, Szocs and Janosi.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
LinkOut - more resources
Full Text Sources

