Interferon-inducible phospholipids govern IFITM3-dependent endosomal antiviral immunity
- PMID: 36970857
- PMCID: PMC10183820
- DOI: 10.15252/embj.2022112234
Interferon-inducible phospholipids govern IFITM3-dependent endosomal antiviral immunity
Abstract
The interferon-induced transmembrane proteins (IFITM) are implicated in several biological processes, including antiviral defense, but their modes of action remain debated. Here, taking advantage of pseudotyped viral entry assays and replicating viruses, we uncover the requirement of host co-factors for endosomal antiviral inhibition through high-throughput proteomics and lipidomics in cellular models of IFITM restriction. Unlike plasma membrane (PM)-localized IFITM restriction that targets infectious SARS-CoV2 and other PM-fusing viral envelopes, inhibition of endosomal viral entry depends on lysines within the conserved IFITM intracellular loop. These residues recruit Phosphatidylinositol 3,4,5-trisphosphate (PIP3) that we show here to be required for endosomal IFITM activity. We identify PIP3 as an interferon-inducible phospholipid that acts as a rheostat for endosomal antiviral immunity. PIP3 levels correlated with the potency of endosomal IFITM restriction and exogenous PIP3 enhanced inhibition of endocytic viruses, including the recent SARS-CoV2 Omicron variant. Together, our results identify PIP3 as a critical regulator of endosomal IFITM restriction linking it to the Pi3K/Akt/mTORC pathway and elucidate cell-compartment-specific antiviral mechanisms with potential relevance for the development of broadly acting antiviral strategies.
Keywords: IFITM; SARS-CoV2; innate immunity; phosphatidylinositol 3,4,5-trisphosphate (PIP3); viral entry.
© 2023 The Authors. Published under the terms of the CC BY NC ND 4.0 license.
Conflict of interest statement
The authors declare that they no conflict of interest.
Figures
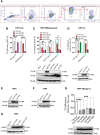
- A
Gating strategy for acquisition of transduction efficiencies in the different cell lines.
- B
THP1 overexpressing (Oe) IFITM3, IFITM1, or control were transduced with VSV‐gp or measles‐gp‐pseudotyped LVs at MOI 1 (mean ± SEM, n > 6 biological replicates run in technical duplicate), transduction efficiency was measured by flow cytometry 5‐day post‐TD as % of BFP+ cells. P‐values are for Mann–Whitney test, ****for P < 0.0001.
- C
THP1 overexpressing IFITM3 wild‐type, IFITM3 PM‐confined mutants or control were transduced at MOI 1 with PM‐fusing LVs (mean ± SEM, n > 5 biological replicates run in technical duplicate), P‐values are for Mann–Whitney test, * for P < 0.05, **for P < 0.01. IFITM1, IFITM2, IFITM3, Y20F, and Y20A overexpression was confirmed by WB in THP1.
- D
THP1 overexpressing the putative IFITM3 isoform (∆21) encoded by the SNP rs‐1225‐C or control were transduced with either VSV‐gp or measles‐gp‐pseudotyped LVs. Transduction efficiency was measured 5 days later (mean ± SEM, n = 5 biological replicates run in technical duplicate), P‐values are for one‐sample t‐test, *** for P < 0.001, **** for P < 0.0001. IFITM3‐WT and IFITM3‐∆21 overexpression was confirmed by WB in THP1.
- E, F
IFITM3 protein expression was evaluated by WB at time of TD in THP1 overexpressing IFITM3, IFITM3‐4KR, or IFITM3‐3KR.
- G
THP1 overexpressing single lysine mutants IFITM3 or control were transduced with VSV‐gp‐pseudotyped LV. Transduction efficiency was measured by flow cytometry 5‐day post‐TD (mean ± SEM, n = 6 biological replicates run in technical duplicate). Overexpression was confirmed by WB at the time of TD. P‐values are for one‐sample t‐test versus Oe‐Luc = 1. *** for P < 0.001, **** for P < 0.0001.
- H
THP1 overexpressing IFITM3, IFITM3‐Y20F, IFITM3‐Y20F‐3KR, or control were validated by WB.
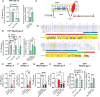
- A, B
THP1 overexpressing IFITM3, IFITM3‐Y20F, IFITM3‐∆21, or control were pretreated or not with Amphotericin B for 1 h and then transduced with a VSV‐gp (A) or measles‐gp (B) pseudotyped LV. Transduction efficiencies were evaluated at FACS 5 days later. Fold rescue of transduction over the DMSO control was calculated. Data are shown as mean ± SEM (n = 5–8 biological replicates run in technical duplicate). P‐values are for one‐sample t‐test versus Oe‐Luc DMSO = 1, ** for P < 0.01, *** for P < 0.001.
- C
Schematic representation of IFITM3 protein. Lysine residues (K) are highlighted in red.
- D
Alignment of IFITM3 orthologs in vertebrates. Conserved lysine residues are marked in yellow.
- E, F
THP1 overexpressing IFITM3, IFITM3 lysine‐less mutant (4KR) (E), IFITM3 CIL‐lysine mutant (3KR) (F), or control were transduced with VSV‐gp‐pseudotyped LV. Transduction efficiencies were evaluated by flow cytometry 5‐day post‐TD. Data represent the mean ± SEM (n = 9–6 biological replicates run in technical duplicate). P‐values are for one‐sample t‐test verso Oe‐Luc DMSO = 1. * for P < 0.05, ** for P < 0.01, **** for P < 0.0001 and ns for not significant.
- G, H
THP1 overexpressing IFITM1, IFITM1‐3KR (G), IFITM3, IFITM3‐Y20F, IFITM3‐Y20F‐3KR (H) or control were transduced with measles‐gp‐pseudotyped LV. Transduction efficiencies were evaluated by flow cytometry 5‐day post‐TD. Data represent the mean ± SEM (n = 4–9 biological replicates run in technical duplicate). P‐values are for one‐sample t‐test verso Oe‐Luc DMSO = 1. * for P < 0.05, ** for P < 0.01, **** for P < 0.0001.
- I
THP1 overexpressing IFITM3‐Y20F‐3KR mutant or control were exposed or not to Amphotericin B for 1 h and then transduced with measles‐gp‐pseudotyped LV. Transduction levels were measured by flow cytometry 5‐day post‐TD (mean ± SEM, n = 8 biological replicates run in technical duplicate). P‐values are for one‐sample t‐test versus Oe‐Luc DMSO = 1 or Mann–Whitney test for Ampho B versus DMSO in Oe‐Y20F‐3KR. * for P < 0.05, ** for P < 0.01.
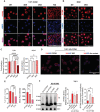
- A, B
Immunofluorescence images were performed using TCS SP5 Leica confocal microscope, 60× with oil on THP1 overexpressing IFITM3‐WT, IFITM3‐4KR, IFITM3‐3KR, or IFITM3‐Y20F (A) (n = 22 images acquired from three biological replicates) and primary monocyte‐derived macrophages (MDM) overexpressing IFITM3‐WT, IFITM3‐4KR, IFITM3‐3KR (B) (n = 12 images acquired from three independent donors). Colocalization (purple areas) of IFITM3 (in red) with the lysosome‐associated membrane protein 1 (LAMP1) marker (in blue) was evaluated. Representative zoomed images are shown, scale bar 100 μm.
- C
Quantification of Pearson's colocalization coefficient was performed on Oe‐THP1 and MDM (n = 22–12 images acquired from three biological replicates).
- D
IFITM3‐IFITM3 interactions (red dots) were evaluated by proximity ligation assay (PLA) in THP1 knockout for endogenous IFITM3 and expressing either HA‐IFITM3 and His‐IFITM3, HA‐3KR and His‐3KR or His‐dimer mutant IFITM3 (D.M) and HA‐dimer mutant. Images were acquired on TCS SP5 Leica confocal microscope 60× with oil. Representative images are shown (n = 8 images of four biological replicates), scale bar 40 μm.
- E
Number of foci and PLA foci intensity were measured on ImageJ and normalized over the number of nuclei (mean ± SEM, n = 4 biological replicates).
- F
IFITM3 dimerization was assessed by native PAGE followed by WB analysis in THP1. IFITM3 higher‐order protein complex bands are highlighted by the asterisk. IFITM3 dimers and higher molecular weight complexes were quantified over the normalizer using ImageJ (mean ± SEM, n = 7 biological replicates). P‐values are for Mann–Whitney test. * for P < 0.05, ** for P < 0.01.
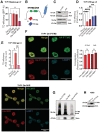
THP1 overexpressing IFITM3, IFITM3‐3KR, IFITM3‐Y20F‐3KR, or control were transduced with measles‐gp‐pseudotyped LV. Transduction efficiency was evaluated 5‐day post‐TD (mean ± SEM, n = 3 biological replicates run in technical duplicate). P‐values are for one paired t‐test. * for P < 0.05.
Schematic representation of IFITM3 wild‐type or 3KR mutant tagged with hemagglutinin (HA) or Histidine at the N terminus (NT).
Overexpression of IFITM3‐tagged constructs was validated in THP1 by WB.
THP1 overexpressing tagged IFITM3, tagged IFITM3‐3KR or control were transduced with VSV‐gp‐pseudotyped LV. Transduction was evaluated 5‐day post‐TD (mean ± SEM, n = 6 biological replicates run in technical duplicate). P‐values are for one‐sample t‐test versus Oe‐Luc = 1 for comparisons with tagged IFITM3 or Mann–Whitney test for comparisons between tagged IFITM3 and tagged IFITM3‐3KR. ** for P < 0.01, **** for P < 0.0001.
THP1 overexpressing tagged IFITM3, tagged IFITM3‐dimer mutant or control were transduced with VSV‐gp‐pseudotyped LV. Transduction was evaluated 5‐day post‐TD (mean ± SEM, n = 6 biological replicates run in technical duplicate). P‐values are for one‐sample t‐test versus Oe‐Luc = 1 for comparisons with tagged IFITM3 or Mann–Whitney test for comparisons between tagged IFITM3 and tagged IFITM3‐dimer mutant. * for P < 0.05, ** for P < 0.01.
HA‐IFITM3, His‐IFITM3, HA‐3KR, His‐3KR, and Lamp1 levels were measured by immunofluorescence (mean ± SEM, n = 3 biological replicates; zoomed images are shown, scale bar 40 μm). The Pearson's correlation coefficient of HA, His, and Lamp1 colocalization of three independent biological experiments was calculated. P‐values are for Mann–Whitney test. ns for not significant.
IFITM3 dimerization was assessed by native PAGE followed by WB analysis in THP1 (n = 4 biological replicates).
IFITM3 overexpression was confirmed in K562 at the time of TD.
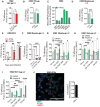
- A
K562 overexpressing IFITM3 or control were transduced with VSV‐gp‐pseudotyped LV (mean ± SEM, n = 6 biological replicates run in technical duplicate). Transduction levels were calculated at FACS 5‐day post‐TD measuring the % of BFP expression within Oe‐cells. Data are represented as Folds over the Oe‐Luc control. P‐values are for one‐sample t‐test versus Oe‐Luc = 1. ns for not significant.
- B
K562 were prestimulated with IFNα and then transduced with VSV‐gp‐pseudotyped LV. Transduction was evaluated 5 days after TD (mean ± SEM, n = 4 biological replicates run in technical duplicate). Data are represented as Folds over the Oe‐Luc control. P‐values are for one‐sample t‐test versus ‐IFNα = 1. ns for not significant.
- C
Interferon‐stimulated genes (ISGs) mRNA expression were measured 24‐h post‐IFNα stimulation by RT–qPCR in K562 (mean ± SEM, n = 3 biological replicates run in technical duplicate).
- D
K562 were prestimulated with IFNα and then transduced with measles‐gp‐pseudotyped LV. Transduction was measured 5‐day post‐TD (mean ± SEM, n = 3 biological replicates run in technical duplicate). P‐values are for one‐sample t‐test versus ‐IFNα = 1. *** for P < 0.001.
- E
K562 were prestimulated or not with IFNα for 24 h and then inoculated with the infectious ZIKV (mean ± SEM, n = 3 biological replicates run in technical triplicates). Viral supernatant was collected at different time points and titered on Vero E6 cells. P‐values are for Mann–Whitney test. ** for P < 0.01, *** for P < 0.001.
- F
K562 overexpressing IFITM1, IFITM1‐3KR, or control were transduced with measles‐gp‐pseudotyped LV. Transduction efficiency was evaluated 5‐day post‐TD (mean ± SEM, n = 4 biological replicates run in technical duplicate). P‐values are for one‐sample t‐test versus Oe‐Luc = 1. **** for P < 0.0001.
- G–I
K562 overexpressing IFITM3, IFITM3‐Y20F, IFITM3‐Y20F‐3KR, or control were pre‐exposed or not to Ampho B and then transduced with measles‐gp (G), BaEV‐gp (H) or RD114‐gp (I) pseudotyped LV. Transduction efficiencies were measured by flow cytometry 5 days after TD (mean ± SEM, n = 5–4‐4 biological replicates run in technical duplicate). P‐values are for one‐sample t‐test versus Oe‐Luc DMSO = 1 or unpaired t‐test for Ampho B versus DMSO. * for P < 0.05, ** for P < 0.01, *** for P < 0.001, **** for P < 0.0001.
- J
IFITM3 localization was analyzed by IF in K562, a representative zoomed image is shown (n = 12 images of three biological replicates), scale bar 100 μm. Pearson's colocalization coefficient was measured with ImageJ.

- A
Venn diagram of wild‐type (WT) IFITM3 (gray marking) and 4KR‐IFITM3 (green marking) interactors identified by mass spectrometry in THP1.
- B
IFITM3‐WT or IFITM3‐4KR were immunoprecipitated in THP1, interactors were identified by mass spectrometry. The heatmap shows proteins with significant higher affinity for IFITM3‐WT compared with IFITM3‐4KR (Benjamini–Hochberg FDR < 0.05). In red interactors linked or localizing in endosomes and lysosomes.
- C
Venn diagram of IFITM3 interactors (gray marking, 74 proteins) ISG interactors (turquoise marking, 1,371 proteins), vORF interactors (yellow marking, 574 proteins), and viral interactors (green marking, 3,902 proteins). The labels indicate the number of proteins in the overlaps of the dataset.
- D
IFITM3 interactors protein map and functionally enriched terms (Benjamini–Hochberg FDR < 0.05). Phosphatidylinositol binding is highlighted in yellow.
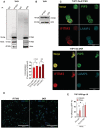
IFITM3 wild‐type or IFITM3‐4KR immunoprecipitations were confirmed by WB.
IFITM3 wild‐type or IFITM3‐3KR expression levels were measured in THP1 KO for endogenous IFITM3.
IFITM3 and PIP3 colocalization with LAMP1 was evaluated by immunofluorescence. Representative zoomed images are shown (mean ± SEM, n = 3 biological replicates), scale bar 40 μm. The Pearson's correlation coefficient of HA, His, and Lamp1 colocalization of three independent biological experiments was calculated. ns for not significant.
Total PIP3 was quantified by IF in THP1 cells overexpressing the double‐tagged His/HA IFITM3 wild‐type or IFITM3‐3KR. Representative images are shown.
THP1 KO for IFITM3 or control were stimulated with IFNα for 24 h and then transduced with VSV‐gp‐pseudotyped LV in the presence or absence of Rapamycin. Transduction levels were measured 5 days after TD (mean ± SEM, n = 6 biological replicates run in technical duplicate). P‐values are for Wilcoxon test, * for P < 0.05.
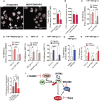
Interaction between IFITM3 wild‐type or 3KR and Phosphatidynositol‐3 phosphate (PIP3; red dots) was evaluated by proximity ligation assay. Images were acquired using TCS SP5 Leica confocal microscope 60× with oil in THP1 knockout for endogenous IFITM3 and expressing either IFITM3 wild‐type or IFITM3‐3KR. Number of foci were counted on ImageJ and normalized over the number of nuclei (mean ± SEM, n = 5 biological replicates). White arrows indicate foci, scale bar 40 μm.
Phosphatidynositol‐3 phosphate (PIP3) levels were measured by immunofluorescence in THP1 overexpressing the His‐IFITM3/HA‐IFITM3 and His‐3KR/HA‐3KR. PIP3 was quantified via ImageJ (mean ± SEM, n = 5 biological replicates).
THP1 overexpressing IFITM3 or control were transduced with VSV‐gp‐pseudotyped LV in the presence or absence of Rapamycin (Rapa). Transduction efficiency was measured at FACS 5‐day post‐TD (mean ± SEM, n = 6 biological replicates run in technical duplicate. P‐values are for one‐sample t‐test versus Oe‐Luc DMSO = 1 and Mann–Whitney test for Rapa versus DMSO. ** for P < 0.01, **** for P < 0.0001.
THP1 overexpressing IFITM3‐Y20F‐3KR or control were exposed or not to Rapamycin and transduced with measles‐gp‐pseudotyped LV. Transduction efficiency was evaluated by flow cytometry 5‐day post‐TD (mean ± SEM, n = 4 biological replicates run in technical duplicate). P‐values are for one‐sample t‐test versus Oe‐Luc DMSO = 1. ** for P < 0.0.
Hematopoietic stem and progenitor cells (HSPC) were transduced with VSV‐gp or measles‐gp‐pseudotyped LV in the presence or absence of Rapamycin. Transduction was evaluated at FACS 5 days later measuring the % of BFP expression (mean ± SEM, n = 4 biological replicates run in technical duplicate). P‐values are for Mann–Whitney test. * for P < 0.05.
THP1 overexpressing IFITM3 or control were transduced with VSV‐gp‐pseudotyped LV in the presence or absence of MK2206. Transduction efficiency was analyzed 5‐day post‐TD (mean ± SEM, n = 6 biological replicates run in technical duplicate). P‐values are for one‐sample t‐test versus Oe‐Luc DMSO = 1 and for Mann–Whitney test for Oe‐IFITM3 DMSO versus MK2206, * for P < 0.05, *** for P < 0.001.
Human hematopoietic stem and progenitor cells were pretreated with MK6602 for 1 h and then transduced with VSV‐gp‐pseudotyped LV (mean ± SEM, n = 4 biological replicates run in technical duplicate). P‐values are for one‐sample t‐test versus Oe‐Luc DMSO = 1. * for P < 0.05.
THP1 overexpressing IFITM3, KO‐IFITM3, or control were transduced with VSV‐gp‐pseudotyped LV after 4 h of pretreatment with LY94002. Transduction efficiency was analyzed 5‐day post‐TD (mean ± SEM, n = 5 biological replicates run in technical duplicate). P‐values are for one‐sample t‐test versus Oe‐Luc DMSO and Mann–Whitney test for IFITM3 DMSO versus IFITM3 LY294002, KO‐IFITM3 DMSO versus KO‐IFITM3 LY294002 as well as for fold rescue of transduction. * for P < 0.05, *** for P < 0.001.
Schematic representation of IFITM3‐PIP3 signaling cascade acting on the PI3K‐mTOR–AKT axes, respectively, blocked by Rapamycin and MK2206.
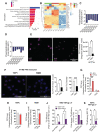
- A
RNA sequencing performed in HSPC (CD34), THP1, and K562. Pathways differentially regulated in HSPC and THP1 in comparison with K562 are highlighted.
- B
Heatmap representation of genes involved in PI3K and phosphatidylinositol phosphate binding pathway. Upregulated genes are shown in red, and downregulated genes are shown in blue.
- C, D
Levels of phosphatidylinositol (PI) precursors of PIP3 and PIP2 were measured through lipidomic analysis in THP1, HSPC, and K562. The results are presented as log plots of the PI ratio in K562 over THP1 (C) or HSPC (D).
- E
PIP3 levels were measured by immunofluorescence (scale bar 40 μm) and quantified as integrated density with ImageJ software in THP1 and K562 overexpressing IFITM3 (mean ± SEM, n = 3 biological replicates). P‐values are for Mann–Whitney test, * for P < 0.05.
- F
Interaction between IFITM3 and Phosphatidynositol‐3 phosphate (PIP3; red dots) was evaluated by proximity ligation assay using TCS SP5 Leica confocal microscope 60× with oil in THP1 or K562 expressing same levels of IFITM3. Number of foci and PLA foci intensity were measured on ImageJ and normalized over the number of nuclei (mean ± SEM, n = 4 biological replicates run in technical duplicate), scale bar 40 μm. P‐values are for Mann–Whitney test, ** for P < 0.01, *** for P < 0.001.).
- G–I
PIP3 levels were measured by immunofluorescence and quantified as integrated density with ImageJ software in THP1 and K562 (G), HSPC (H), and MDM (I) prestimulated or not with IFNα (mean ± SEM, n = 3 biological replicates. P–values are for Mann–Whitney test, *** for P < 0.001.
- J
PIP3 coupled to a specific PIP carrier (PIP3 + Carr Ctrl) or Carr Ctrl alone were delivered to K562 overexpressing IFITM3, control or KO for IFITM3. All conditions were transduced with VSV‐gp‐pseudotyped LV. Transduction levels were evaluated 5‐day post‐TD (mean ± SEM, n = 5 biological replicates run in technical duplicate). P‐values are for one‐sample t‐test versus Ctrl Carr = 1. * for P < 0.05, ** for P < 0.01. The effect on transduction of exogenous PIP3 in K562 is represented as Fold versus Carr Ctrl.
- K
HSPC pretreated with PIP3 coupled to a specific PIP carrier (PIP3 + Carr Ctrl) or Carr Ctrl alone were transduced with VSV‐gp‐pseudotyped LV. Transduction levels were evaluated 5‐day post‐TD measuring the % of BFP expression (mean ± SEM, n = 5 biological replicates run in technical duplicate). P‐values are for Mann–Whitney test, * for P < 0.05.
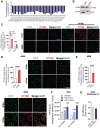
- A
All Phosphatidylinositol (PI) species were measured in K562 and THP1. Log plots of PI ratio in K562 versus THP1 are shown.
- B
IFITM3 expression was evaluated by WB in THP1 and K562 cells KO for endogenous IFITM3 and overexpressing IFITM3.
- C–E
PIP3 levels were measured by immunofluorescence in THP1 and K562 (C), HSPC (D) and MDM (E) prestimulated with IFNα. (mean ± SEM, n = 3 biological replicates). P‐values are for Mann–Whitney test, * for P < 0.05. Representative images are shown, scale bar 100 μm.
- F
K562 were provided of exogenous PIP3 or Carr Ctrl alone. Successful PIP3 intake was verified by IF 5 and 15‐min postdelivery by IF (n = 8 acquired from two independent biological experiments).
- G
K562 were provided of exogenous PIP3 or Carr Ctrl alone and then transduced with measles‐gp‐pseudotyped LV. Transduction was measured 5 days later (mean ± SEM, n = 4 biological replicates run in duplicate).

- A
Calu3 overexpressing IFITM1, IFITM2, IFITM3, PM‐mutant IFITM3 Y20A, or control were transduced with SARS‐CoV2 Spike‐pseudotyped LV (mean ± SEM, n > 6 biological replicates run in technical duplicate). Transduction efficiency was measured by FACS 5 days after transduction, P‐values are for one‐sample paired t‐test versus Oe‐Luc Ctrl = 1, **for P < 0.01.
- B, C
Calu3 cells overexpressing IFITM1, IFITM2, IFITM3, IFITM3 Y20A, or control were infected with SARS‐CoV2. Viral titer was calculated by plaque assay in Vero E6 cells (mean ± SEM, n = 4 biological replicates run in technical duplicate). P‐values are for Mann–Whitney test, * for P < 0.05, *** for P < 0.001.
- D
Calu3 overexpressing IFITM3‐Δ21 or control were infected with SARS CoV2 for 3 days. Viral titer was assessed by plaque assay in Vero E6 cells (mean ± SEM, n = 3 biological replicates run in technical duplicate). P‐values are for Mann–Whitney test, * for P < 0.05.
- E
Calu3 and THP1 were infected with SARS‐CoV2 at MOI 1 and 0.01 respectively. Viral supernatant was titered on Vero E6 cells (mean ± SEM, n = 4 biological replicates run in technical triplicate).
- F
THP1 cells overexpressing IFITM1, IFITM2, IFITM3, IFITM3 plasma membrane‐confined mutants or control were infected with SARS‐CoV2 at MOI 0.01. Titers were calculated by tittering viral supernatants on Vero E6 cells (mean ± SEM, n = 4 biological replicates run in technical triplicate). P‐values are for Mann–Whitney test, * for P < 0.05.
- G
Calu3 overexpressing IFITM1, IFITM3, or control were challenged to SARS‐CoV2 Omicron‐pseudotyped LV. Transduction efficiency was evaluated 5‐day post‐transduction (mean ± SEM, n = 7 biological replicates run in technical duplicate). P‐values are for one‐sample t‐test versus Oe‐Luc Ctrl = 1, **for P < 0.01, ****for P < 0.0001.
- H, I
Calu3 cells overexpressing IFITM1, IFITM2, IFITM3, or control (H) or IFITM3, Y20A, or control (I) were infected with Omicron at MOI 1. Viral supernatants were collected at different time points and tittered on Vero E6 cells. Data are presented as mean ± SEM (n = 4–4 biological replicates run in technical triplicate). P‐values are for Mann–Whitney test, **for P < 0.01, ***for P < 0.001, ****for P < 0.0001.
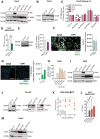
- A
Overexpression of IFITM1, IFITM2, IFITM3, and IFITM3‐Y20A was verified by WB in Calu3.
- B
IFITM3‐WT and IFITM3‐∆21 overexpression was confirmed by WB in Calu3.
- C
Calu3 overexpressing IFITM1, IFITM2, IFITM3, IFITM3‐Y20A mutant, and control were transduced with LV‐pseudotyped with different Spike variants. Error bars are presented as mean ± SEM (n = 6 biological replicates run in duplicate); P‐values are for one‐sample t‐test, * for P < 0.05, **for P < 0.01, ***for P < 0.001.
- D
THP1 overexpressing ∆21‐IFITM3 or control were infected with SARS CoV2 for 3 days. Viral titer was assessed by plaque assay in Vero E6 cells (mean ± SEM, n = 4 biological replicates run in technical triplicate). P‐values are for Mann–Whitney test, ** for P < 0.01.
- E
ACE2 protein level was measured in Calu2 and THP1. Quantification was performed by ImageJ over the normalizer (mean ± SEM, n = 5 biological replicates).
- F, G
Human Ace2 (F) and TMPRSS2 (G) expression at the cell surface was analyzed in Calu3 and THP1 cells by immunofluorescence. Protein quantification was performed using ImageJ software (mean ± SEM, n = 4–3 biological replicates); representative images are shown (n = 12–6 images), scale bar 100 μm.
- H
A549 overexpressing ACE2 was validated by RT–qPCR. (mean ± SEM, n = 3 biological replicates).
- I, J
Overexpression of IFITM1, IFITM2, or IFITM3 was measured in A549 (I) or Vero E6 cells (J).
- K
A549 cells overexpressing IFITM1, IFITM2, IFITM3, or control were infected with SARS‐CoV2 at MOI 1. Viral supernatants were collected at different time points and tittered on Vero E6 cells. Data are presented as mean ± SEM (n = 4 biological replicates run in technical duplicate). P‐values are for Mann–Whitney test, *P < 0.05.
- L
Vero E6 cells overexpressing IFITM1, IFITM2, IFITM3, or control were infected with SARS‐CoV2 at MOI 1. Titers were calculated through direct plaque assay 72‐h postinfection. (n = 3 biological replicates run in technical duplicate). P‐values are for Mann–Whitney test, *P < 0.05.
- M
Calu3 overexpressing IFITM3, IFITM3‐Y20F, IFITM3‐Y20F‐3KR, or control was validated by WB.
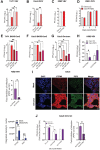
- A, B
VSV (A) or ZIKV (B) viral supernatants were collected 1‐day postinfection in THP1 (A) or Calu3 (B) overexpressing IFITM3, IFITM3 lysine‐less mutant (4KR), IFITM3 CIL‐lysine mutant (3KR) or control and titered on VERO E6 cells. Data represent the mean ± SEM (n = 5–3 biological replicates run in technical duplicate). P‐values are for Mann–Whitney test. * for P < 0.05, ** for P < 0.01.
- C, D
K562 were inoculated with the infectious VSV (C) or ZIKV (D) (mean ± SEM, n = 3 biological replicates run in technical duplicate). Viral supernatant was collected at different time points and titered on Vero E6 cells.
- E–G
THP1 (E) or Calu3 (F, G) overexpressing IFITM3, IFITM3‐Y20F mutant, IFITM3‐Y20F‐3KR mutant or control were inoculated with SARS‐CoV2 (E, F) or Omicron (G). Viral titers were measured on Vero E6 cells. Data are presented as mean ± SEM (n = 4–4‐3 biological replicates run in technical duplicate). P‐values are for Mann–Whitney test, * for P < 0.05, ** for P < 0.01, ns for not significant.
- H
K562 overexpressing IFITM3 or KO for IFITM3 were pretreated with PIP3 coupled to a specific PIP carrier (PIP3 + Carr Ctrl) or Carr Ctrl and then infected with VSV. Infectious supernatant were collected after 3 days and tittered in Vero E6 cells (mean ± SEM, n = 6 biological replicates run in technical duplicate). P‐values are for Wilcoxon signed‐ranked test, * for P < 0.05. The inhibitory effect of exogenous PIP3 on infection is represented as Fold versus Carr Ctrl (mean ± SEM, n = 6 biological replicates run in technical duplicate). P‐values are for Mann–Whitney test, * for P < 0.05.
- I
IFITM3 and PIP3 levels were measured by immunofluorescence (scale bar 100 μm) and quantified as integrated density with ImageJ software in Calu3 infected or not with Omicron (n = 3 biological replicates). P‐values are for Mann–Whitney test, ** for P < 0.01.
- J
PIP3 coupled to a specific PIP carrier (PIP3 + Carr Ctrl) or Carr Ctrl were provided to Calu3 overexpressing IFITM3, control or KO for IFITM3 prior to infection with SARS‐CoV2 Omicron. Infectious supernatants were collected after 3 days and tittered in Vero E6 cells (mean ± SEM, n = 4 biological replicates run in technical duplicate). P‐values are for Wilcoxon signed‐ranked test, * for P < 0.05. The inhibitory effect of exogenous PIP3 on infection is represented as Fold versus Carr Ctrl (mean ± SEM, n = 4 biological replicates run in technical duplicate). P‐values are for Mann–Whitney test, * for P < 0.05.
References
-
- Aleem A, Akbar Samad AB, Slenker AK (2022) Emerging variants of SARS‐CoV‐2 and novel therapeutics against coronavirus (COVID‐19). Treasure Island, FL: StatPearls; - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

