Extrachromosomal Amplification of Human Papillomavirus Episomes Is a Mechanism of Cervical Carcinogenesis
- PMID: 36971511
- PMCID: PMC10239328
- DOI: 10.1158/0008-5472.CAN-22-3030
Extrachromosomal Amplification of Human Papillomavirus Episomes Is a Mechanism of Cervical Carcinogenesis
Abstract
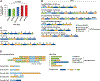
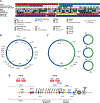
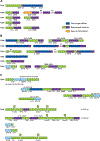
Multimers of the HPV genome are generated in cervical tumors replicating as extrachromosomal episomes, which is associated with deletion and rearrangement of the HPV genome and provides a mechanism for oncogenesis without integration.
©2023 American Association for Cancer Research.
Conflict of interest statement
Conflicts of interest
The authors declare no potential conflicts of interest.
Figures




References
-
- Schiffman M, Doorbar J, Wentzensen N, de Sanjose S, Fakhry C, Monk BJ, et al. Carcinogenic human papillomavirus infection. Nat Rev Dis Primers 2016;2:16086. - PubMed
-
- Walboomers JM, Jacobs MV, Manos MM, Bosch FX, Kummer JA, Shah KV, et al. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol 1999;189:12–9 - PubMed
-
- Schiffman M, Castle PE. The promise of global cervical-cancer prevention. N Engl J Med 2005;353:2101–4 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

