Scaffold-Scaffold Interaction Facilitates Cell Polarity Development in Caulobacter crescentus
- PMID: 36971555
- PMCID: PMC10127582
- DOI: 10.1128/mbio.03218-22
Scaffold-Scaffold Interaction Facilitates Cell Polarity Development in Caulobacter crescentus
Abstract
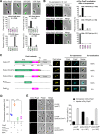
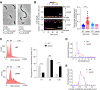
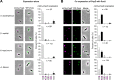
Cell polarity development is the prerequisite for cell differentiation and generating biodiversity. In the model bacterium Caulobacter crescentus, the polarization of the scaffold protein PopZ during the predivisional cell stage plays a central role in asymmetric cell division. However, our understanding of the spatiotemporal regulation of PopZ localization remains incomplete. In the current study, a direct interaction between PopZ and the new pole scaffold PodJ is revealed, which plays a primary role in triggering the new pole accumulation of PopZ. The coiled-coil 4-6 domain in PodJ is responsible for interacting with PopZ in vitro and promoting PopZ transition from monopolar to bipolar in vivo. Elimination of the PodJ-PopZ interaction impairs the PopZ-mediated chromosome segregation by affecting both the positioning and partitioning of the ParB-parS centromere. Further analyses of PodJ and PopZ from other bacterial species indicate this scaffold-scaffold interaction may represent a widespread strategy for spatiotemporal regulation of cell polarity in bacteria. IMPORTANCE Caulobacter crescentus is a well-established bacterial model to study asymmetric cell division for decades. During cell development, the polarization of scaffold protein PopZ from monopolar to bipolar plays a central role in C. crescentus asymmetric cell division. Nevertheless, the spatiotemporal regulation of PopZ has remained unclear. Here, we demonstrate that the new pole scaffold PodJ functions as a regulator in triggering PopZ bipolarization. The primary regulatory role of PodJ was demonstrated in parallel by comparing it with other known PopZ regulators, such as ZitP and TipN. Physical interaction between PopZ and PodJ ensures the timely accumulation of PopZ at the new cell pole and the inheritance of the polarity axis. Disruption of the PodJ-PopZ interaction impaired PopZ-mediated chromosome segregation and may lead to a decoupling of DNA replication from cell division during the cell cycle. Together, the scaffold-scaffold interaction may provide an underlying infrastructure for cell polarity development and asymmetric cell division.
Keywords: Caulobacter crescentus; PodJ; PopZ; asymmetric cell division; cell polarity; chromosome segregation; scaffold proteins.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
