The small GTPase Rit2 modulates LRRK2 kinase activity, is required for lysosomal function and protects against alpha-synuclein neuropathology
- PMID: 36973269
- PMCID: PMC10042831
- DOI: 10.1038/s41531-023-00484-2
The small GTPase Rit2 modulates LRRK2 kinase activity, is required for lysosomal function and protects against alpha-synuclein neuropathology
Abstract
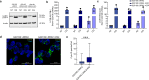
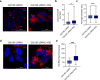
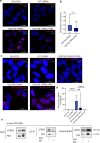
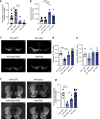
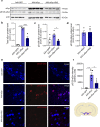
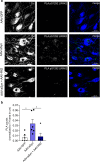
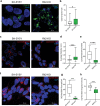
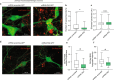
In Parkinson's disease (PD) misfolded alpha-synuclein (aSyn) accumulates in the substantia nigra, where dopaminergic neurons are progressively lost. The mechanisms underlying aSyn pathology are still unclear, but they are hypothesized to involve the autophagy-lysosome pathway (ALP). LRRK2 mutations are a major cause of familial and sporadic PD, and LRRK2 kinase activity has been shown to be involved in pS129-aSyn inclusion modulation. We observed selective downregulation of the novel PD risk factor RIT2 in vitro and in vivo. Rit2 overexpression in G2019S-LRRK2 cells rescued ALP abnormalities and diminished aSyn inclusions. In vivo, viral mediated overexpression of Rit2 operated neuroprotection against AAV-A53T-aSyn. Furthermore, Rit2 overexpression prevented the A53T-aSyn-dependent increase of LRRK2 kinase activity in vivo. On the other hand, reduction of Rit2 levels leads to defects in the ALP, similar to those induced by the G2019S-LRRK2 mutation. Our data indicate that Rit2 is required for correct lysosome function, inhibits overactive LRRK2 to ameliorate ALP impairment, and counteracts aSyn aggregation and related deficits. Targeting Rit2 could represent an effective strategy to combat neuropathology in familial and idiopathic PD.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources

