Soybean gene co-expression network analysis identifies two co-regulated gene modules associated with nodule formation and development
- PMID: 36975024
- PMCID: PMC10189762
- DOI: 10.1111/mpp.13327
Soybean gene co-expression network analysis identifies two co-regulated gene modules associated with nodule formation and development
Abstract
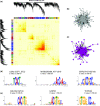
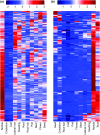
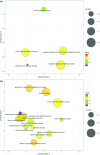
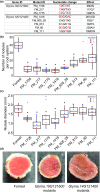
Gene co-expression network analysis is an efficient systems biology approach for the discovery of novel gene functions and trait-associated gene modules. To identify clusters of functionally related genes involved in soybean nodule formation and development, we performed a weighted gene co-expression network analysis. Two nodule-specific modules (NSM-1 and NSM-2, containing 304 and 203 genes, respectively) were identified. The NSM-1 gene promoters were significantly enriched in cis-binding elements for ERF, MYB, and C2H2-type zinc transcription factors, whereas NSM-2 gene promoters were enriched in cis-binding elements for TCP, bZIP, and bHLH transcription factors, suggesting a role of these regulatory factors in the transcriptional activation of nodule co-expressed genes. The co-expressed gene modules included genes with potential novel roles in nodulation, including those involved in xylem development, transmembrane transport, the ethylene signalling pathway, cytoskeleton organization, cytokinesis and regulation of the cell cycle, regulation of meristem initiation and growth, transcriptional regulation, DNA methylation, and histone modifications. Functional analysis of two co-expressed genes using TILLING mutants provided novel insight into the involvement of unsaturated fatty acid biosynthesis and folate metabolism in nodule formation and development. The identified gene co-expression modules provide valuable resources for further functional genomics studies to dissect the genetic basis of nodule formation and development in soybean.
Keywords: TILLING; gene co-expression network; nodule formation and development; soybean.
© 2023 The Authors. Molecular Plant Pathology published by British Society for Plant Pathology and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
Similar articles
-
Large-scale analysis of putative soybean regulatory gene expression identifies a Myb gene involved in soybean nodule development.Plant Physiol. 2009 Nov;151(3):1207-20. doi: 10.1104/pp.109.144030. Epub 2009 Sep 15. Plant Physiol. 2009. PMID: 19755542 Free PMC article.
-
Co-regulatory network analysis of the main secondary metabolite (SM) biosynthesis in Crocus sativus L.Sci Rep. 2024 Jul 9;14(1):15839. doi: 10.1038/s41598-024-65870-z. Sci Rep. 2024. PMID: 38982154 Free PMC article.
-
Whole genome co-expression analysis of soybean cytochrome P450 genes identifies nodulation-specific P450 monooxygenases.BMC Plant Biol. 2010 Nov 9;10:243. doi: 10.1186/1471-2229-10-243. BMC Plant Biol. 2010. PMID: 21062474 Free PMC article.
-
Exploring the complexity of soybean (Glycine max) transcriptional regulation using global gene co-expression networks.Planta. 2020 Nov 16;252(6):104. doi: 10.1007/s00425-020-03499-8. Planta. 2020. PMID: 33196909
-
Systematic identification of functional modules and cis-regulatory elements in Arabidopsis thaliana.BMC Bioinformatics. 2011 Nov 24;12 Suppl 12(Suppl 12):S2. doi: 10.1186/1471-2105-12-S12-S2. BMC Bioinformatics. 2011. PMID: 22168340 Free PMC article.
Cited by
-
Overexpression of soybean GmNAC19 and GmGRAB1 enhances root growth and water-deficit stress tolerance in soybean.Front Plant Sci. 2023 May 31;14:1186292. doi: 10.3389/fpls.2023.1186292. eCollection 2023. Front Plant Sci. 2023. PMID: 37324708 Free PMC article.
-
Differential symbiotic compatibilities between rhizobium strains and cultivated and wild soybeans revealed by anatomical and transcriptome analyses.Front Plant Sci. 2024 Sep 3;15:1435632. doi: 10.3389/fpls.2024.1435632. eCollection 2024. Front Plant Sci. 2024. PMID: 39290740 Free PMC article.
-
Deep Learning Model for Classifying and Evaluating Soybean Leaf Disease Damage.Int J Mol Sci. 2023 Dec 20;25(1):106. doi: 10.3390/ijms25010106. Int J Mol Sci. 2023. PMID: 38203277 Free PMC article.
-
The analysis of the genetic loci affecting phenotypic plasticity of soybean isoflavone content by dQTG.seq model.Theor Appl Genet. 2024 Dec 17;138(1):9. doi: 10.1007/s00122-024-04798-4. Theor Appl Genet. 2024. PMID: 39688708
References
-
- Banuelos, J. , Martínez‐Romero, E. , Montaño, N.M. & Camargo‐Ricalde, S.L. (2021) Folates in legume root nodules. Physiologia Plantarum, 171, 447–452. - PubMed
-
- Brechenimacher, L. , Kim, M.Y. , Benitez, M. , Li, M. , Joshi, T. , Calla, B. et al. (2008) Transcription profiling of soybean nodulation by Bradyrhizobium japonicum . Molecular Plant‐Microbe Interactions, 21, 631–645. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

