Genomic Analysis of Vancomycin-Resistant Staphylococcus aureus Isolates from the 3rd Case Identified in the United States Reveals Chromosomal Integration of the vanA Locus
- PMID: 36975781
- PMCID: PMC10100801
- DOI: 10.1128/spectrum.04317-22
Genomic Analysis of Vancomycin-Resistant Staphylococcus aureus Isolates from the 3rd Case Identified in the United States Reveals Chromosomal Integration of the vanA Locus
Abstract
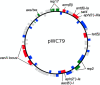
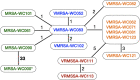
Vancomycin-resistant Staphylococcus aureus (VRSA) is a human pathogen of significant public health concern. Although the genome sequences of individual VRSA isolates have been published over the years, very little is known about the genetic changes of VRSA within a patient over time. A total of 11 VRSA, 3 vancomycin-resistant enterococci (VRE), and 4 methicillin-resistant S. aureus (MRSA) isolates, collected over a period of 4.5 months in 2004 from a patient in a long-term-care facility in New York State, were sequenced. A combination of long- and short-read sequencing technologies was used to obtain closed assemblies for chromosomes and plasmids. Our results indicate that a VRSA isolate emerged as the result of the transfer of a multidrug resistance plasmid from a coinfecting VRE to an MRSA isolate. The plasmid then integrated into the chromosome via homologous recombination mediated between two regions derived from remnants of transposon Tn5405. Once integrated, the plasmid underwent further reorganization in one isolate, while two others lost the staphylococcal cassette chromosome mec element (SCCmec) determinant that confers methicillin-resistance. The results presented here explain how a few recombination events can lead to multiple pulsed-field gel electrophoresis (PFGE) patterns that could be mistaken for vastly different strains. A vanA gene cluster that is located on a multidrug resistance plasmid that is integrated into the chromosome could result in the continuous propagation of resistance, even in the absence of selective pressure from antibiotics. The genome comparison presented here sheds light on the emergence and evolution of VRSA within a single patient that will enhance our understanding VRSA genetics. IMPORTANCE High-level vancomycin-resistant Staphylococcus aureus (VRSA) began to emerge in the United States in 2002 and has since then been reported worldwide. Our study reports the closed genome sequences of multiple VRSA isolates obtained in 2004 from a single patient in New York State. Our results show that the vanA resistance locus is located on a mosaic plasmid that confers resistance to multiple antibiotics. In some isolates, this plasmid integrated into the chromosome via homologous recombination between two ant(6)-sat4-aph(3') antibiotic resistance loci. This is, to our knowledge, the first report of a chromosomal vanA locus in VRSA; the effect of this integration event on MIC values and plasmid stability in the absence of antibiotic selection remains poorly understood. These findings highlight the need for a better understanding of the genetics of the vanA locus and plasmid maintenance in S. aureus to address the increase of vancomycin resistance in the health care setting.
Keywords: Staphylococcus aureus; VRSA; antibiotic resistance; vancomycin; vancomycin-resistant Staphylococcus aureus; whole-genome sequencing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Bugg TD, Wright GD, Dutka-Malen S, Arthur M, Courvalin P, Walsh CT. 1991. Molecular basis for vancomycin resistance in Enterococcus faecium BM4147: biosynthesis of a depsipeptide peptidoglycan precursor by vancomycin resistance proteins VanH and VanA. Biochemistry 30:10408–10415. doi: 10.1021/bi00107a007. - DOI - PubMed
LinkOut - more resources
Full Text Sources

