Diversity of the Antimicrobial Peptide Genes in Collembola
- PMID: 36975900
- PMCID: PMC10051947
- DOI: 10.3390/insects14030215
Diversity of the Antimicrobial Peptide Genes in Collembola
Abstract
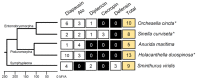
Multidrug-resistant bacteria are a current health crisis threatening the world's population, and scientists are looking for new drugs to combat them. Antimicrobial peptides (AMPs), which are part of the organism's innate immune system, are a promising new drug class as they can disrupt bacterial cell membranes. This study explored antimicrobial peptide genes in collembola, a non-insect hexapod lineage that has survived in microbe-rich habitats for millions of years, and their antimicrobial peptides have not been thoroughly investigated. We used in silico analysis (homology-based gene identification, physicochemical and antimicrobial property prediction) to identify AMP genes from the genomes and transcriptomes of five collembola representing three main suborders: Entomobryomorpha (Orchesella cincta, Sinella curviseta), Poduromorpha (Holacanthella duospinosa, Anurida maritima), and Symphypleona (Sminthurus viridis). We identified 45 genes belonging to five AMP families, including (a) cysteine-rich peptides: diapausin, defensin, and Alo; (b) linear α-helical peptide without cysteine: cecropin; (c) glycine-rich peptide: diptericin. Frequent gene gains and losses were observed in their evolution. Based on the functions of their orthologs in insects, these AMPs potentially have broad activity against bacteria, fungi, and viruses. This study provides candidate collembolan AMPs for further functional analysis that could lead to medicinal use.
Keywords: AMP evolution; AMP gene identification; antimicrobial peptide; collembola immunity; drug discovery.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures







Similar articles
-
Insect antimicrobial peptides and their applications.Appl Microbiol Biotechnol. 2014 Jul;98(13):5807-22. doi: 10.1007/s00253-014-5792-6. Epub 2014 May 9. Appl Microbiol Biotechnol. 2014. PMID: 24811407 Free PMC article. Review.
-
Analysis of the genome of the New Zealand giant collembolan (Holacanthella duospinosa) sheds light on hexapod evolution.BMC Genomics. 2017 Oct 17;18(1):795. doi: 10.1186/s12864-017-4197-1. BMC Genomics. 2017. PMID: 29041914 Free PMC article.
-
Screening and Functional Analyses of Novel Cecropins from Insect Transcriptome.Insects. 2023 Sep 29;14(10):794. doi: 10.3390/insects14100794. Insects. 2023. PMID: 37887806 Free PMC article.
-
Antimicrobial peptide-like genes in Nasonia vitripennis: a genomic perspective.BMC Genomics. 2010 Mar 19;11:187. doi: 10.1186/1471-2164-11-187. BMC Genomics. 2010. PMID: 20302637 Free PMC article.
-
Antimicrobial peptides: the ancient arm of the human immune system.Virulence. 2010 Sep-Oct;1(5):440-64. doi: 10.4161/viru.1.5.12983. Virulence. 2010. PMID: 21178486 Review.
Cited by
-
Diversity and Molecular Evolution of Antimicrobial Peptides in Caecilian Amphibians.Toxins (Basel). 2024 Mar 14;16(3):150. doi: 10.3390/toxins16030150. Toxins (Basel). 2024. PMID: 38535816 Free PMC article.
References
-
- O’Neill J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. Government of the United Kingdom; London, UK: 2016. [(accessed on 10 December 2022)]. Available online: https://wellcomecollection.org/works/thvwsuba.
-
- Talebi Bezmin Abadi A., Rizvanov A.A., Haertlé T., Blatt N.L. World Health Organization Report: Current Crisis of Antibiotic Resistance. BioNanoScience. 2019;9:778–788. doi: 10.1007/s12668-019-00658-4. - DOI
LinkOut - more resources
Full Text Sources

