POLQ inhibition elicits an immune response in homologous recombination-deficient pancreatic adenocarcinoma via cGAS/STING signaling
- PMID: 36976649
- PMCID: PMC10232002
- DOI: 10.1172/JCI165934
POLQ inhibition elicits an immune response in homologous recombination-deficient pancreatic adenocarcinoma via cGAS/STING signaling
Abstract
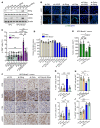
Pancreatic ductal adenocarcinoma (PDAC) is a highly lethal malignancy that harbors mutations in homologous recombination-repair (HR-repair) proteins in 20%-25% of cases. Defects in HR impart a specific vulnerability to poly ADP ribose polymerase inhibitors and platinum-containing chemotherapy in tumor cells. However, not all patients who receive these therapies respond, and many who initially respond ultimately develop resistance. Inactivation of the HR pathway is associated with the overexpression of polymerase theta (Polθ, or POLQ). This key enzyme regulates the microhomology-mediated end-joining (MMEJ) pathway of double-strand break (DSB) repair. Using human and murine HR-deficient PDAC models, we found that POLQ knockdown is synthetically lethal in combination with mutations in HR genes such as BRCA1 and BRCA2 and the DNA damage repair gene ATM. Further, POLQ knockdown enhances cytosolic micronuclei formation and activates signaling of cyclic GMP-AMP synthase-stimulator of interferon genes (cGAS-STING), leading to enhanced infiltration of activated CD8+ T cells in BRCA2-deficient PDAC tumors in vivo. Overall, POLQ, a key mediator in the MMEJ pathway, is critical for DSB repair in BRCA2-deficient PDAC. Its inhibition represents a synthetic lethal approach to blocking tumor growth while concurrently activating the cGAS-STING signaling pathway to enhance tumor immune infiltration, highlighting what we believe to be a new role for POLQ in the tumor immune environment.
Keywords: Cancer; Cell Biology; DNA repair; T cells.
Figures

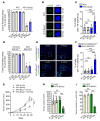
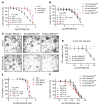
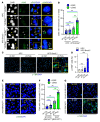
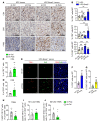
Comment in
- Polymerase theta inhibition steps on the cGAS pedal doi: 10.1172/JCI170660
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

