Target Class Profiling of Small-Molecule Methyltransferases
- PMID: 36976909
- PMCID: PMC10983791
- DOI: 10.1021/acschembio.3c00124
Target Class Profiling of Small-Molecule Methyltransferases
Abstract
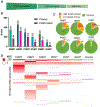
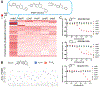
Target class profiling (TCP) is a chemical biology approach to investigate understudied biological target classes. TCP is achieved by developing a generalizable assay platform and screening curated compound libraries to interrogate the chemical biological space of members of an enzyme family. In this work, we took a TCP approach to investigate inhibitory activity across a set of small-molecule methyltransferases (SMMTases), a subclass of methyltransferase enzymes, with the goal of creating a launchpad to explore this largely understudied target class. Using the representative enzymes nicotinamide N-methyltransferase (NNMT), phenylethanolamine N-methyltransferase (PNMT), histamine N-methyltransferase (HNMT), glycine N-methyltransferase (GNMT), catechol O-methyltransferase (COMT), and guanidinoacetate N-methyltransferase (GAMT), we optimized high-throughput screening (HTS)-amenable assays to screen 27,574 unique small molecules against all targets. From this data set, we identified a novel inhibitor which selectively inhibits the SMMTase HNMT and demonstrated how this platform approach can be leveraged for a targeted drug discovery campaign using the example of HNMT.
Conflict of interest statement
The authors declare no competing financial interests. Data for all assays are publicly available on PubChem. AID #s: 1745853 (Primary screen), 1745852 (confirmation screen), 1745851 (counter screen)
Figures






References
-
- Copeland RA; Solomon ME; Richon VM, Protein methyltransferases as a target class for drug discovery. Nat Rev Drug Discov 2009, 8 (9), 724–32. - PubMed
-
- Heilker R; Wolff M; Tautermann CS; Bieler M, G-protein-coupled receptor-focused drug discovery using a target class platform approach. Drug Discov Today 2009, 14 (5–6), 231–40. - PubMed
-
- Carter AJ; Kraemer O; Zwick M; Mueller-Fahrnow A; Arrowsmith CH; Edwards AM, Target 2035: probing the human proteome. Drug Discov Today 2019, 24 (11), 2111–2115. - PubMed
-
- Posy SL; Hermsmeier MA; Vaccaro W; Ott KH; Todderud G; Lippy JS; Trainor GL; Loughney DA; Johnson SR, Trends in kinase selectivity: insights for target class-focused library screening. J Med Chem 2011, 54 (1), 54–66. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

