Structural features of the Notch ankyrin domain-Deltex WWE2 domain heterodimer determined by NMR spectroscopy and functional implications
- PMID: 36977409
- PMCID: PMC10338078
- DOI: 10.1016/j.str.2023.03.003
Structural features of the Notch ankyrin domain-Deltex WWE2 domain heterodimer determined by NMR spectroscopy and functional implications
Abstract
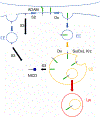
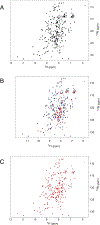
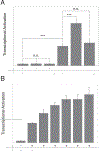
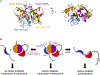
The Notch signaling pathway, an important cell fate determination pathway, is modulated by the ubiquitin ligase Deltex. Here, we investigate the structural basis for Deltex-Notch interaction. We used nuclear magnetic resonance (NMR) spectroscopy to assign the backbone of the Drosophila Deltex WWE2 domain and mapped the binding site of the Notch ankyrin (ANK) domain to the N-terminal WWEA motif. Using cultured Drosophila S2R+ cells, we find that point substitutions within the ANK-binding surface of Deltex disrupt Deltex-mediated enhancement of Notch transcriptional activation and disrupt ANK binding in cells and in vitro. Likewise, ANK substitutions that disrupt Notch-Deltex heterodimer formation in vitro block disrupt Deltex-mediated stimulation of Notch transcription activation and diminish interaction with full-length Deltex in cells. Surprisingly, the Deltex-Notch intracellular domain (NICD) interaction is not disrupted by deletion of the Deltex WWE2 domain, suggesting a secondary Notch-Deltex interaction. These results show the importance of the WWEA:ANK interaction in enhancing Notch signaling.
Keywords: Deltex; NMR; Notch; Notch signaling; RING domain; WWE(2) motifs; ankyrin repeats; protein-protein interactions; ubiquitin ligase.
Copyright © 2023 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

