Late-Stage Metastatic Melanoma Emerges through a Diversity of Evolutionary Pathways
- PMID: 36977461
- PMCID: PMC10236155
- DOI: 10.1158/2159-8290.CD-22-1427
Late-Stage Metastatic Melanoma Emerges through a Diversity of Evolutionary Pathways
Abstract
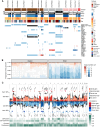
Understanding the evolutionary pathways to metastasis and resistance to immune-checkpoint inhibitors (ICI) in melanoma is critical for improving outcomes. Here, we present the most comprehensive intrapatient metastatic melanoma dataset assembled to date as part of the Posthumous Evaluation of Advanced Cancer Environment (PEACE) research autopsy program, including 222 exome sequencing, 493 panel-sequenced, 161 RNA sequencing, and 22 single-cell whole-genome sequencing samples from 14 ICI-treated patients. We observed frequent whole-genome doubling and widespread loss of heterozygosity, often involving antigen-presentation machinery. We found KIT extrachromosomal DNA may have contributed to the lack of response to KIT inhibitors of a KIT-driven melanoma. At the lesion-level, MYC amplifications were enriched in ICI nonresponders. Single-cell sequencing revealed polyclonal seeding of metastases originating from clones with different ploidy in one patient. Finally, we observed that brain metastases that diverged early in molecular evolution emerge late in disease. Overall, our study illustrates the diverse evolutionary landscape of advanced melanoma.
Significance: Despite treatment advances, melanoma remains a deadly disease at stage IV. Through research autopsy and dense sampling of metastases combined with extensive multiomic profiling, our study elucidates the many mechanisms that melanomas use to evade treatment and the immune system, whether through mutations, widespread copy-number alterations, or extrachromosomal DNA. See related commentary by Shain, p. 1294. This article is highlighted in the In This Issue feature, p. 1275.
©2023 The Authors; Published by the American Association for Cancer Research.
Figures
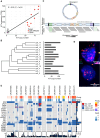
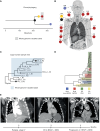
![Figure 3. Late-emerging brain metastases have a lower copy-number burden. A, wGII per metastatic site. Site-specific null distributions of mean wGII were generated by randomizing sample sets (from any metastatic site) while keeping patient contributions constant (see Methods). **, P ≤ 0.01. Leptomen., leptomeninges. B, Correlation between brain copy-number (CN) distance to other sites and time of emergence of brain metastases after stage IV diagnosis in days. C, Growth dynamics of tumors in patient CRUKP5107. The brain lesion (in orange) was detected in only the last two scans after the targeted therapy [BRAF inhibitor (i) + MEKi], ICI (PD-1i + CTLA4i), and chemotherapy courses. PD, progressive disease; PR, partial response; SD, stable disease. D, SNV and indel phylogenetic tree of tumor clones in patient CRUKP5107. E, The mutational signature contributions to each clone in the phylogeny in D are shown. MMR, mismatch repair. F, The anatomic distribution of clones. Each pie chart represents a sample with its clonal composition indicated by the colors. A multiregional sampling of the same tumor is indicated by the gray dashed lines. BR, brain; LI, liver; LU, lung; PC, pericardium; ST, soft tissue.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/b29b/10236155/a28f49b8e5e6/1364fig3.gif)
Comment in
-
Melanoma Genomics: Shifting Focus from Inter- to Intrapatient Variation.Cancer Discov. 2023 Jun 2;13(6):1294-1296. doi: 10.1158/2159-8290.CD-23-0340. Cancer Discov. 2023. PMID: 37264823
References
-
- Robert C, Grob JJ, Stroyakovskiy D, Karaszewska B, Hauschild A, Levchenko E, et al. Five-year outcomes with dabrafenib plus trametinib in metastatic melanoma. N Engl J Med 2019;381:626–36. - PubMed
-
- Larkin J, Chiarion-Sileni V, Gonzalez R, Grob JJ, Rutkowski P, Lao CD, et al. Five-year survival with combined nivolumab and ipilimumab in advanced melanoma. N Engl J Med 2019;381:1535–46. - PubMed
-
- Hayward NK, Wilmott JS, Waddell N, Johansson PA, Field MA, Nones K, et al. Whole-genome landscapes of major melanoma subtypes. Nature 2017;545:175–80. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 21999/CRUK_/Cancer Research UK/United Kingdom
- C11496/A17786/CRUK_/Cancer Research UK/United Kingdom
- 17786/CRUK_/Cancer Research UK/United Kingdom
- 30025/CRUK_/Cancer Research UK/United Kingdom
- U01 CA247439/CA/NCI NIH HHS/United States
- MR/P014712/1/MRC_/Medical Research Council/United Kingdom
- C11496/A30025/CRUK_/Cancer Research UK/United Kingdom
- FC10988/MRC_/Medical Research Council/United Kingdom
- MR/V033077/1/MRC_/Medical Research Council/United Kingdom
- A29911/CRUK_/Cancer Research UK/United Kingdom
- FC10988/CRUK_/Cancer Research UK/United Kingdom
- FC10988/WT_/Wellcome Trust/United Kingdom
- 29911/CRUK_/Cancer Research UK/United Kingdom
- FC001169/WT_/Wellcome Trust/United Kingdom
- C69256/A30194/CRUK_/Cancer Research UK/United Kingdom
LinkOut - more resources
Full Text Sources
Medical

