FBXO11 governs macrophage cell death and inflammation in response to bacterial toxins
- PMID: 36977592
- PMCID: PMC10053445
- DOI: 10.26508/lsa.202201735
FBXO11 governs macrophage cell death and inflammation in response to bacterial toxins
Abstract
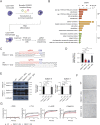
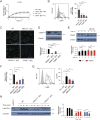
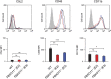
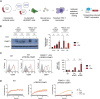
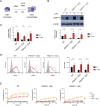
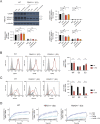
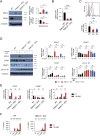
Staphylococcus aureus causes severe infections such as pneumonia and sepsis depending on the pore-forming toxin Panton-Valentine leukocidin (PVL). PVL kills and induces inflammation in macrophages and other myeloid cells by interacting with the human cell surface receptor, complement 5a receptor 1 (C5aR1). C5aR1 expression is tighly regulated and may thus modulate PVL activity, although the mechanisms involved remain incompletely understood. Here, we used a genome-wide CRISPR/Cas9 screen and identified F-box protein 11 (FBXO11), an E3 ubiquitin ligase complex member, to promote PVL toxicity. Genetic deletion of FBXO11 reduced the expression of C5aR1 at the mRNA level, whereas ectopic expression of C5aR1 in FBXO11-/- macrophages, or priming with LPS, restored C5aR1 expression and thereby PVL toxicity. In addition to promoting PVL-mediated killing, FBXO11 dampens secretion of IL-1β after NLRP3 activation in response to bacterial toxins by reducing mRNA levels in a BCL-6-dependent and BCL-6-independent manner. Overall, these findings highlight that FBXO11 regulates C5aR1 and IL-1β expression and controls macrophage cell death and inflammation following PVL exposure.
© 2023 Jeon et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
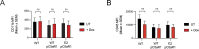

References
-
- Baron BW, Nucifora G, McCabe N, Espinosa R 3rd, Le Beau MM, McKeithan TW (1993) Identification of the gene associated with the recurring chromosomal translocations t(3;14)(q27;q32) and t(3;22)(q27;q11) in B-cell lymphomas. Proc Natl Acad Sci U S A 90: 5262–5266. 10.1073/pnas.90.11.5262 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
