Lonafarnib and everolimus reduce pathology in iPSC-derived tissue engineered blood vessel model of Hutchinson-Gilford Progeria Syndrome
- PMID: 36977745
- PMCID: PMC10050176
- DOI: 10.1038/s41598-023-32035-3
Lonafarnib and everolimus reduce pathology in iPSC-derived tissue engineered blood vessel model of Hutchinson-Gilford Progeria Syndrome
Abstract
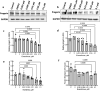
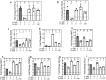
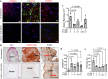
Hutchinson-Gilford Progeria Syndrome (HGPS) is a rare, fatal genetic disease that accelerates atherosclerosis. With a limited pool of HGPS patients, clinical trials face unique challenges and require reliable preclinical testing. We previously reported a 3D tissue engineered blood vessel (TEBV) microphysiological system fabricated with iPSC-derived vascular cells from HGPS patients. HGPS TEBVs exhibit features of HGPS atherosclerosis including loss of smooth muscle cells, reduced vasoactivity, excess extracellular matrix (ECM) deposition, inflammatory marker expression, and calcification. We tested the effects of HGPS therapeutics Lonafarnib and Everolimus separately and together, currently in Phase I/II clinical trial, on HGPS TEBVs. Everolimus decreased reactive oxygen species levels, increased proliferation, reduced DNA damage in HGPS vascular cells, and improved vasoconstriction in HGPS TEBVs. Lonafarnib improved shear stress response of HGPS iPSC-derived endothelial cells (viECs) and reduced ECM deposition, inflammation, and calcification in HGPS TEBVs. Combination treatment with Lonafarnib and Everolimus produced additional benefits such as improved endothelial and smooth muscle marker expression and reduced apoptosis, as well as increased TEBV vasoconstriction and vasodilation. These results suggest that a combined trial of both drugs may provide cardiovascular benefits beyond Lonafarnib, if the Everolimus dose can be tolerated.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Gordon, L. B., Brown, W. T. & Collins, F. S. in GeneReviews® (eds M. Adam, P. et al.) (University of Washington, 2019).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

