Transformation and expressional studies of GaZnF gene to improve drought tolerance in Gossypium hirsutum
- PMID: 36977831
- PMCID: PMC10050179
- DOI: 10.1038/s41598-023-32383-0
Transformation and expressional studies of GaZnF gene to improve drought tolerance in Gossypium hirsutum
Abstract
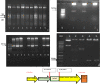
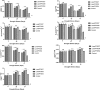
Drought stress is the major limiting factor in plant growth and production. Cotton is a significant crop as textile fiber and oilseed, but its production is generally affected by drought stress, mainly in dry regions. This study aimed to investigate the expression of Zinc finger transcription factor's gene (GaZnF) to enhance the drought tolerance in Gossypium hirsutum. Sequence features of the GaZnF protein were recognized through different bioinformatics tools like multiple sequence alignment analysis, phylogenetic tree for evolutionary relationships, Protein motifs, a transmembrane domain, secondary structure and physio-chemical properties indicating that GaZnF is a stable protein. CIM-482, a local Gossypium hirsutum variety was transformed with GaZnF through Agrobacterium-mediated transformation method with 2.57% transformation efficiency. The integration of GaZnF was confirmed through Southern blot showing 531 bp, and Western blot indicated a 95 kDa transgene-GUS fusion band in transgenic plants. The normalized real-time expression analysis revealed the highest relative fold spatial expression of cDNA of GaZnF within leaf tissues at vegetative and flowering stages under drought stress. Morphological, physiological and biochemical parameters of transgenic cotton plants at 05- and 10-day drought stress was higher than those of non-transgenic control plants. The values of fresh and dry biomass, chlorophyll content, photosynthesis, transpiration rate, and stomatal conductance reduced in GaZnF transgenic cotton plants at 05- and 10-day drought stress, but their values were less low in transgenic plants than those of non-transgenic control plants. These findings showed that GaZnF gene expression in transgenic plants could be a valuable source for the development of drought-tolerant homozygous lines through breeding.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
-
- Costa, M.-C. D. & Farrant, J. M. Vol. 8 553 (MDPI, 2019).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

