GVC: efficient random access compression for gene sequence variations
- PMID: 36978010
- PMCID: PMC10044409
- DOI: 10.1186/s12859-023-05240-0
GVC: efficient random access compression for gene sequence variations
Abstract
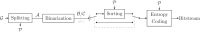
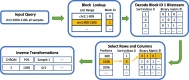
Background: In recent years, advances in high-throughput sequencing technologies have enabled the use of genomic information in many fields, such as precision medicine, oncology, and food quality control. The amount of genomic data being generated is growing rapidly and is expected to soon surpass the amount of video data. The majority of sequencing experiments, such as genome-wide association studies, have the goal of identifying variations in the gene sequence to better understand phenotypic variations. We present a novel approach for compressing gene sequence variations with random access capability: the Genomic Variant Codec (GVC). We use techniques such as binarization, joint row- and column-wise sorting of blocks of variations, as well as the image compression standard JBIG for efficient entropy coding.
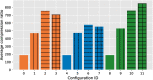
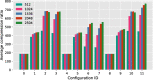
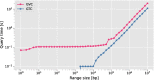
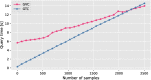
Results: Our results show that GVC provides the best trade-off between compression and random access compared to the state of the art: it reduces the genotype information size from 758 GiB down to 890 MiB on the publicly available 1000 Genomes Project (phase 3) data, which is 21% less than the state of the art in random-access capable methods.
Conclusions: By providing the best results in terms of combined random access and compression, GVC facilitates the efficient storage of large collections of gene sequence variations. In particular, the random access capability of GVC enables seamless remote data access and application integration. The software is open source and available at https://github.com/sXperfect/gvc/ .
Keywords: Compression; Random access; VCF; Variants.
© 2023. The Author(s).
Conflict of interest statement
JV, CR, VT, YGA, and JO have filed the patent application DE102021100199A1, which covers parts of the methods presented in the manuscript.
Figures
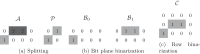
References
MeSH terms
LinkOut - more resources
Full Text Sources

