Clinical Resistant Strains of Enterococci and Their Correlation to Reduced Susceptibility to Biocides: Phenotypic and Genotypic Analysis of Macrolides, Lincosamides, and Streptogramins
- PMID: 36978327
- PMCID: PMC10044631
- DOI: 10.3390/antibiotics12030461
Clinical Resistant Strains of Enterococci and Their Correlation to Reduced Susceptibility to Biocides: Phenotypic and Genotypic Analysis of Macrolides, Lincosamides, and Streptogramins
Abstract
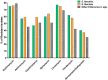
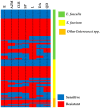
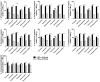
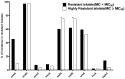
Enterococci are troublesome nosocomial, opportunistic Gram-positive cocci bacteria showing enhanced resistance to many commonly used antibiotics. This study aims to investigate the prevalence and genetic basis of antibiotic resistance to macrolides, lincosamides, and streptogramins (MLS) in Enterococci, as well as the correlation between MLS resistance and biocide resistance. From 913 clinical isolates collected from King Khalid Hospital, Hail, Saudi Arabia, 131 isolates were identified as Enterococci spp. The susceptibility of the clinical enterococcal isolates to several MLS antibiotics was determined, and the resistance phenotype was detected by the triple disk method. The MLS-involved resistance genes were screened in the resistant isolates. The current results showed high resistance rates to MLS antibiotics, and the constitutive resistance to all MLS (cMLS) was the most prevalent phenotype, observed in 76.8% of resistant isolates. By screening the MLS resistance-encoding genes in the resistant isolates, the erythromycin ribosome methylase (erm) genes that are responsible for methylation of bacterial 23S rRNA were the most detected genes, in particular, ermB. The ereA esterase-encoding gene was the most detected MLS modifying-encoding genes, more than lnuA (adenylation) and mphC (phosphorylation). The minimum inhibitory concentrations (MICs) of commonly used biocides were detected in resistant isolates and correlated with the MICs of MLS antibiotics. The present findings showed a significant correlation between MLS resistance and reduced susceptibility to biocides. In compliance with the high incidence of the efflux-encoding genes, especially mefA and mefE genes in the tolerant isolates with higher MICs to both MLS antibiotics and biocides, the efflux of resistant isolates was quantified, and there was a significant increase in the efflux of resistant isolates with higher MICs as compared to those with lower MICs. This could explain the crucial role of efflux in developing cross-resistance to both MLS antibiotics and biocides.
Keywords: Enterococci; Enterococci faecalis; Enterococci faecium; MLS phenotypes; biocides; genotyping; lincosamides; macrolides; streptogramins.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Kirschner C., Maquelin K., Pina P., Ngo Thi N.A., Choo-Smith L.P., Sockalingum G.D., Sandt C., Ami D., Orsini F., Doglia S.M., et al. Classification and identification of enterococci: A comparative phenotypic, genotypic, and vibrational spectroscopic study. J. Clin. Microbiol. 2001;39:1763–1770. doi: 10.1128/JCM.39.5.1763-1770.2001. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

