Computer-Aided Drug Design and Synthesis of Rhenium Clotrimazole Antimicrobial Agents
- PMID: 36978486
- PMCID: PMC10044843
- DOI: 10.3390/antibiotics12030619
Computer-Aided Drug Design and Synthesis of Rhenium Clotrimazole Antimicrobial Agents
Abstract
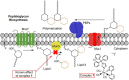
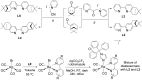
In the context of the global health issue caused by the growing occurrence of antimicrobial resistance (AMR), the need for novel antimicrobial agents is becoming alarming. Inorganic and organometallic complexes represent a relatively untapped source of antibiotics. Here, we report a computer-aided drug design (CADD) based on a 'scaffold-hopping' approach for the synthesis and antibacterial evaluation of fac-Re(I) tricarbonyl complexes bearing clotrimazole (ctz) as a monodentate ligand. The prepared molecules were selected following a pre-screening in silico analysis according to modification of the 2,2'-bipyridine (bpy) ligand in the coordination sphere of the complexes. CADD pointed to chiral 4,5-pinene and 5,6-pinene bipyridine derivatives as the most promising candidates. The corresponding complexes were synthesized, tested toward methicillin-sensitive and -resistant S. aureus strains, and the obtained results evaluated with regard to their binding affinity with a homology model of the S. aureus MurG enzyme. Overall, the title species revealed very similar minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) values as those of the reference compound used as the scaffold in our approach. The obtained docking scores advocate the viability of 'scaffold-hopping' for de novo design, a potential strategy for more cost- and time-efficient discovery of new antibiotics.
Keywords: Staphylococcus aureus MurG; clotrimazole; docking; rhenium.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







References
-
- Jim O.N. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. Government of the United Kingdom; London, UK: 2016.
-
- Claudel M., Schwarte J., Fromm K. New Antimicrobial Strategies Based on Metal Complexes. Chemistry. 2020;2:849–899. doi: 10.3390/chemistry2040056. - DOI
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

