Over-Expressed GATA-1S, the Short Isoform of the Hematopoietic Transcriptional Factor GATA-1, Inhibits Ferroptosis in K562 Myeloid Leukemia Cells by Preventing Lipid Peroxidation
- PMID: 36978786
- PMCID: PMC10045147
- DOI: 10.3390/antiox12030537
Over-Expressed GATA-1S, the Short Isoform of the Hematopoietic Transcriptional Factor GATA-1, Inhibits Ferroptosis in K562 Myeloid Leukemia Cells by Preventing Lipid Peroxidation
Abstract
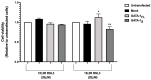
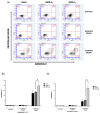
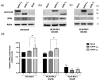
Ferroptosis is a recently recognized form of regulated cell death involving lipid peroxidation. Glutathione peroxidase 4 (GPX4) plays a central role in the regulation of ferroptosis through the suppression of lipid peroxidation generation. Connections have been reported between ferroptosis, lipid metabolism, cancer onset, and drug resistance. Recently, interest has grown in ferroptosis induction as a potential strategy to overcome drug resistance in hematological malignancies. GATA-1 is a key transcriptional factor controlling hematopoiesis-related gene expression. Two GATA-1 isoforms, the full-length protein (GATA-1FL) and a shorter isoform (GATA-1S), are described. A balanced GATA-1FL/GATA-1S ratio helps to control hematopoiesis, with GATA-1S overexpression being associated with hematological malignancies by promoting proliferation and survival pathways in hematopoietic precursors. Recently, optical techniques allowed us to highlight different lipid profiles associated with the expression of GATA-1 isoforms, thus raising the hypothesis that ferroptosis-regulated processes could be involved. Lipidomic and functional analysis were conducted to elucidate these mechanisms. Studies on lipid peroxidation production, cell viability, cell death, and gene expression were used to evaluate the impact of GPX4 inhibition. Here, we provide the first evidence that over-expressed GATA-1S prevents K562 myeloid leukemia cells from lipid peroxidation-induced ferroptosis. Targeting ferroptosis is a promising strategy to overcome chemoresistance. Therefore, our results could provide novel potential therapeutic approaches and targets to overcome drug resistance in hematological malignancies.
Keywords: GATA-1 isoforms; RLS3; cell death; ferroptosis; glutathione peroxidase 4; lipid peroxidation; myeloid leukemia.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. S.T. was supported by a “Nuove emergenze in sicurezza alimentare: strategie per la riduzione del rischio e per la valorizzazione delle produzioni animali- SAFORISK” postdoctoral fellowship.
Figures




References
Grants and funding
LinkOut - more resources
Full Text Sources

