On the Advent of Super-Resolution Microscopy in the Realm of Polycomb Proteins
- PMID: 36979066
- PMCID: PMC10044799
- DOI: 10.3390/biology12030374
On the Advent of Super-Resolution Microscopy in the Realm of Polycomb Proteins
Abstract
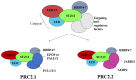
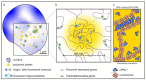
The genomes of metazoans are organized at multiple spatial scales, ranging from the double helix of DNA to whole chromosomes. The intermediate genomic scale of kilobases to megabases, which corresponds to the 50-300 nm spatial scale, is particularly interesting, as the 3D arrangement of chromatin is implicated in multiple regulatory mechanisms. In this context, polycomb group (PcG) proteins stand as major epigenetic modulators of chromatin function, acting prevalently as repressors of gene transcription by combining chemical modifications of target histones with physical crosslinking of distal genomic regions and phase separation. The recent development of super-resolution microscopy (SRM) has strongly contributed to improving our comprehension of several aspects of nano-/mesoscale (10-200 nm) chromatin domains. Here, we review the current state-of-the-art SRM applied to PcG proteins, showing that the application of SRM to PcG activity and organization is still quite limited and mainly focused on the 3D assembly of PcG-controlled genomic loci. In this context, SRM approaches have mostly been applied to multilabel fluorescence in situ hybridization (FISH). However, SRM data have complemented the maps obtained from chromosome capture experiments and have opened a new window to observe how 3D chromatin topology is modulated by PcGs.
Keywords: 3D-SIM; FISH; PRC1; PRC2; STORM; Xist RNA; chromatin organization; oligopaint; polycomb proteins; super-resolution microscopy.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures




References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

