Advances in Mass Spectrometry-Based Single Cell Analysis
- PMID: 36979087
- PMCID: PMC10045136
- DOI: 10.3390/biology12030395
Advances in Mass Spectrometry-Based Single Cell Analysis
Abstract
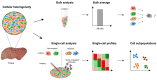
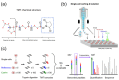
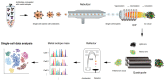
Technological developments and improvements in single-cell isolation and analytical platforms allow for advanced molecular profiling at the single-cell level, which reveals cell-to-cell variation within the admixture cells in complex biological or clinical systems. This helps to understand the cellular heterogeneity of normal or diseased tissues and organs. However, most studies focused on the analysis of nucleic acids (e.g., DNA and RNA) and mass spectrometry (MS)-based analysis for proteins and metabolites of a single cell lagged until recently. Undoubtedly, MS-based single-cell analysis will provide a deeper insight into cellular mechanisms related to health and disease. This review summarizes recent advances in MS-based single-cell analysis methods and their applications in biology and medicine.
Keywords: mass spectrometry; mass spectrometry imaging; metabolomics; proteomics; single-cell analysis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Buettner F., Natarajan K.N., Casale F.P., Proserpio V., Scialdone A., Theis F.J., Teichmann S.A., Marioni J.C., Stegle O. Computational analysis of cell-to-cell heterogeneity in single-cell RNA-sequencing data reveals hidden subpopulations of cells. Nat. Biotechnol. 2015;33:155–160. doi: 10.1038/nbt.3102. - DOI - PubMed
-
- Larsson L.-I. Immunocytochemistry: Theory and Practice. CRC Press; Boca Raton, FL, USA: 1988.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

