Generation of High-Value Genomic Resource in Rice: A "Subgenomic Library" of Low-Light Tolerant Rice Cultivar Swarnaprabha
- PMID: 36979120
- PMCID: PMC10044706
- DOI: 10.3390/biology12030428
Generation of High-Value Genomic Resource in Rice: A "Subgenomic Library" of Low-Light Tolerant Rice Cultivar Swarnaprabha
Abstract
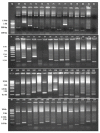
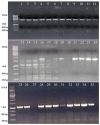
Rice is the major staple food crop for more than 50% of the world's total population, and its production is of immense importance for global food security. As a photophilic plant, its yield is governed by the quality and duration of light. Like all photosynthesizing plants, rice perceives the changes in the intensity of environmental light using phytochromes as photoreceptors, and it initiates a morphological response that is termed as the shade-avoidance response (SAR). Phytochromes (PHYs) are the most important photoreceptor family, and they are primarily responsible for the absorption of the red (R) and far-red (FR) spectra of light. In our endeavor, we identified the morphological differences between two contrasting cultivars of rice: IR-64 (low-light susceptible) and Swarnaprabha (low-light tolerant), and we observed the phenological differences in their growth in response to the reduced light conditions. In order to create genomic resources for low-light tolerant rice, we constructed a subgenomic library of Swarnaprabha that expedited our efforts to isolate light-responsive photoreceptors. The titer of the library was found to be 3.22 × 105 cfu/mL, and the constructed library comprised clones of 4-9 kb in length. The library was found to be highly efficient as per the number of recombinant clones. The subgenomic library will serve as a genomic resource for the Gramineae community to isolate photoreceptors and other genes from rice.
Keywords: Gramineae; far-red light; light-responsive genes; photoreceptors; phytochromes; rice.
Conflict of interest statement
The authors declare no conflict of interest.
Figures








References
-
- Bastia R., Pandit E., Sanghamitra P., Barik S.R., Nayak D.K., Sahoo A., Moharana A., Meher J., Dash P.K., Raj R., et al. Association Mapping for Quantitative Trait Loci Controlling Superoxide Dismutase, Flavonoids, Anthocyanins, Carotenoids, γ-Oryzanol and Antioxidant Activity in Rice. Agronomy. 2022;12:3036. doi: 10.3390/agronomy12123036. - DOI
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

