Orthoparamyxovirinae C Proteins Have a Common Origin and a Common Structural Organization
- PMID: 36979390
- PMCID: PMC10046310
- DOI: 10.3390/biom13030455
Orthoparamyxovirinae C Proteins Have a Common Origin and a Common Structural Organization
Abstract
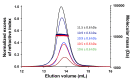
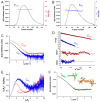
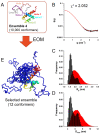
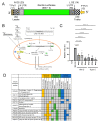
The protein C is a small viral protein encoded in an overlapping frame of the P gene in the subfamily Orthoparamyxovirinae. This protein, expressed by alternative translation initiation, is a virulence factor that regulates viral transcription, replication, and production of defective interfering RNA, interferes with the host-cell innate immunity systems and supports the assembly of viral particles and budding. We expressed and purified full-length and an N-terminally truncated C protein from Tupaia paramyxovirus (TupV) C protein (genus Narmovirus). We solved the crystal structure of the C-terminal part of TupV C protein at a resolution of 2.4 Å and found that it is structurally similar to Sendai virus C protein, suggesting that despite undetectable sequence conservation, these proteins are homologous. We characterized both truncated and full-length proteins by SEC-MALLS and SEC-SAXS and described their solution structures by ensemble models. We established a mini-replicon assay for the related Nipah virus (NiV) and showed that TupV C inhibited the expression of NiV minigenome in a concentration-dependent manner as efficiently as the NiV C protein. A previous study found that the Orthoparamyxovirinae C proteins form two clusters without detectable sequence similarity, raising the question of whether they were homologous or instead had originated independently. Since TupV C and SeV C are representatives of these two clusters, our discovery that they have a similar structure indicates that all Orthoparamyxovirine C proteins are homologous. Our results also imply that, strikingly, a STAT1-binding site is encoded by exactly the same RNA region of the P/C gene across Paramyxovirinae, but in different reading frames (P or C), depending on which cluster they belong to.
Keywords: Paramyxoviridae; overlapping genes; protein structure; viral evolution; virulence factor.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures





References
-
- Lamb R.A., Parks G.D. In: Paramyxoviridae: The Viruses and Their Replication. Fields B.N., Knipe D.M., Howler P.M., editors. Volume 1. Lippincott Williams & Wilkins; Philadelphia, PA, USA: 2013. pp. 957–995.
-
- Vanmechelen B., Meurs S., Horemans M., Loosen A., Maes T.J., Laenen L., Vergote V., Koundouno F.R., Magassouba N., Konde M.K., et al. The Characterization of Multiple Novel Paramyxoviruses Highlights the Diverse Nature of the Subfamily Orthoparamyxovirinae. Virus Evol. 2022;8:veac061. doi: 10.1093/ve/veac061. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

