Structural Insights into the Distortion of the Ribosomal Small Subunit at Different Magnesium Concentrations
- PMID: 36979501
- PMCID: PMC10046523
- DOI: 10.3390/biom13030566
Structural Insights into the Distortion of the Ribosomal Small Subunit at Different Magnesium Concentrations
Abstract
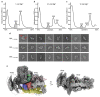
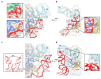
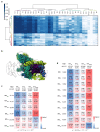
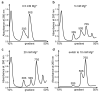
Magnesium ions are abundant and play indispensable functions in the ribosome. A decrease in Mg2+ concentration causes 70S ribosome dissociation and subsequent unfolding. Structural distortion at low Mg2+ concentrations has been observed in an immature pre50S, while the structural changes in mature subunits have not yet been studied. Here, we purified the 30S subunits of E. coli cells under various Mg2+ concentrations and analyzed their structural distortion by cryo-electron microscopy. Upon systematically interrogating the structural heterogeneity within the 1 mM Mg2+ dataset, we observed 30S particles with different levels of structural distortion in the decoding center, h17, and the 30S head. Our model showed that, when the Mg2+ concentration decreases, the decoding center distorts, starting from h44 and followed by the shifting of h18 and h27, as well as the dissociation of ribosomal protein S12. Mg2+ deficiency also eliminates the interactions between h17, h10, h15, and S16, resulting in the movement of h17 towards the tip of h6. More flexible structures were observed in the 30S head and platform, showing high variability in these regions. In summary, the structures resolved here showed several prominent distortion events in the decoding center and h17. The requirement for Mg2+ in ribosomes suggests that the conformational changes reported here are likely shared due to a lack of cellular Mg2+ in all domains of life.
Keywords: CryoEM; magnesium concentration; ribosome; structural distortion.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Alternative conformations and motions adopted by 30S ribosomal subunits visualized by cryo-electron microscopy.RNA. 2020 Dec;26(12):2017-2030. doi: 10.1261/rna.075846.120. Epub 2020 Sep 28. RNA. 2020. PMID: 32989043 Free PMC article.
-
RbfA Is Involved in Two Important Stages of 30S Subunit Assembly: Formation of the Central Pseudoknot and Docking of Helix 44 to the Decoding Center.Int J Mol Sci. 2021 Jun 7;22(11):6140. doi: 10.3390/ijms22116140. Int J Mol Sci. 2021. PMID: 34200244 Free PMC article.
-
[Nature of the heterogeneity of the 30S ribosomal subunits in vitro. II. Two types of inactivation of the 30S subunits of Escherichia coli ribosomes].Mol Biol (Mosk). 1979 Jul-Aug;13(4):752-60. Mol Biol (Mosk). 1979. PMID: 381895 Russian.
-
Chain initiation factor 3 crosslinks to E. coli 30S and 50S ribosomal subunits and alters the UV absorbance spectrum of 70S ribosomes.Nucleic Acids Res. 1982 Sep 25;10(18):5681-93. doi: 10.1093/nar/10.18.5681. Nucleic Acids Res. 1982. PMID: 6755396 Free PMC article.
-
Structural dynamics of ribosomal RNA during decoding on the ribosome.Biochimie. 2002 Aug;84(8):745-54. doi: 10.1016/s0300-9084(02)01409-8. Biochimie. 2002. PMID: 12457562 Review.
Cited by
-
Membraneless protocell confined by a heat flow.Nat Phys. 2025;21(8):1303-1310. doi: 10.1038/s41567-025-02935-4. Epub 2025 Jun 26. Nat Phys. 2025. PMID: 40814307 Free PMC article.
-
Co-purification of the GroEL chaperone during outer membrane vesicle purification: insights from Aeromonas salmonicida subsp. salmonicida.Microbiology (Reading). 2025 Apr;171(4):001558. doi: 10.1099/mic.0.001558. Microbiology (Reading). 2025. PMID: 40293439 Free PMC article.
-
Principles of ion binding to RNA inferred from the analysis of a 1.55 Å resolution bacterial ribosome structure - Part I: Mg2.Nucleic Acids Res. 2025 Jan 7;53(1):gkae1148. doi: 10.1093/nar/gkae1148. Nucleic Acids Res. 2025. PMID: 39791453 Free PMC article.
-
The power of magnesium: unlocking the potential for increased yield, quality, and stress tolerance of horticultural crops.Front Plant Sci. 2023 Oct 24;14:1285512. doi: 10.3389/fpls.2023.1285512. eCollection 2023. Front Plant Sci. 2023. PMID: 37941670 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

