Domain Architecture of the Nonreceptor Tyrosine Kinase Ack1
- PMID: 36980241
- PMCID: PMC10047419
- DOI: 10.3390/cells12060900
Domain Architecture of the Nonreceptor Tyrosine Kinase Ack1
Abstract
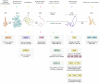
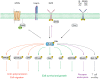
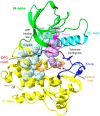
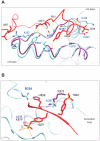
The nonreceptor tyrosine kinase (NRTK) Ack1 comprises a distinct arrangement of non-catalytic modules. Its SH3 domain has a C-terminal to the kinase domain (SH1), in contrast to the typical SH3-SH2-SH1 layout in NRTKs. The Ack1 is the only protein that shares a region of high homology to the tumor suppressor protein Mig6, a modulator of EGFR. The vertebrate Acks make up the only tyrosine kinase (TK) family known to carry a UBA domain. The GTPase binding and SAM domains are also uncommon in the NRTKs. In addition to being a downstream effector of receptor tyrosine kinases (RTKs) and integrins, Ack1 can act as an epigenetic regulator, modulate the degradation of the epidermal growth factor receptor (EGFR), confer drug resistance, and mediate the progression of hormone-sensitive tumors. In this review, we discuss the domain architecture of Ack1 in relation to other protein kinases that possess such defined regulatory domains.
Keywords: Ack1; SAM domain; activated Cdc42-associated kinase; nonreceptor tyrosine kinase; ubiquitin-associated domain.
Conflict of interest statement
The authors declare no conflict of interest.
Figures











References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

