Replication of Real-World Evidence in Oncology Using Electronic Health Record Data Extracted by Machine Learning
- PMID: 36980739
- PMCID: PMC10046618
- DOI: 10.3390/cancers15061853
Replication of Real-World Evidence in Oncology Using Electronic Health Record Data Extracted by Machine Learning
Abstract
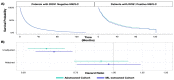
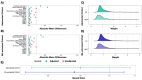
Meaningful real-world evidence (RWE) generation requires unstructured data found in electronic health records (EHRs) which are often missing from administrative claims; however, obtaining relevant data from unstructured EHR sources is resource-intensive. In response, researchers are using natural language processing (NLP) with machine learning (ML) techniques (i.e., ML extraction) to extract real-world data (RWD) at scale. This study assessed the quality and fitness-for-use of EHR-derived oncology data curated using NLP with ML as compared to the reference standard of expert abstraction. Using a sample of 186,313 patients with lung cancer from a nationwide EHR-derived de-identified database, we performed a series of replication analyses demonstrating some common analyses conducted in retrospective observational research with complex EHR-derived data to generate evidence. Eligible patients were selected into biomarker- and treatment-defined cohorts, first with expert-abstracted then with ML-extracted data. We utilized the biomarker- and treatment-defined cohorts to perform analyses related to biomarker-associated survival and treatment comparative effectiveness, respectively. Across all analyses, the results differed by less than 8% between the data curation methods, and similar conclusions were reached. These results highlight that high-performance ML-extracted variables trained on expert-abstracted data can achieve similar results as when using abstracted data, unlocking the ability to perform oncology research at scale.
Keywords: artificial intelligence; cancer; electronic health records; machine learning; natural language processing; oncology; quality; real-world data; real-world evidence.
Conflict of interest statement
At the time of the study, all authors report employment at Flatiron Health, Inc., an independent subsidiary of the Roche Group, and stock ownership in Roche. ME and AC report equity ownership in Flatiron Health, Inc. (initiated before acquisition by Roche in April 2018).
Figures


References
-
- Stark P. Congressional intent for the HITECH Act. [(accessed on 12 January 2023)];Am. J. Manag. Care. 2010 16:SP24–SP28. Available online: https://www.ncbi.nlm.nih.gov/pubmed/21314216. - PubMed
-
- Stewart M., Norden A.D., Dreyer N., Henk H.J., Abernethy A.P., Chrischilles E., Kushi L., Mansfield A.S., Khozin S., Sharon E., et al. An Exploratory Analysis of Real-World End Points for Assessing Outcomes Among Immunotherapy-Treated Patients with Advanced Non–Small-Cell Lung Cancer. JCO Clin. Cancer Inform. 2019;3:1–15. doi: 10.1200/CCI.18.00155. - DOI - PMC - PubMed
-
- Birnbaum B., Nussbaum N., Seidl-Rathkopf K., Agrawal M., Estevez M., Estola E., Haimson J., He L., Larson P., Richardson P. Model-assisted cohort selection with bias analysis for generating large-scale cohorts from the EHR for oncology research. arXiv. 2020 doi: 10.48550/arXiv.2001.09765.2001.09765 - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

