The Molecular and Cellular Basis of Hutchinson-Gilford Progeria Syndrome and Potential Treatments
- PMID: 36980874
- PMCID: PMC10048386
- DOI: 10.3390/genes14030602
The Molecular and Cellular Basis of Hutchinson-Gilford Progeria Syndrome and Potential Treatments
Abstract
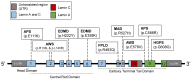
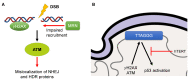
Hutchinson-Gilford progeria syndrome (HGPS) is a rare, autosomal-dominant, and fatal premature aging syndrome. HGPS is most often derived from a de novo point mutation in the LMNA gene, which results in an alternative splicing defect and the generation of the mutant protein, progerin. Progerin behaves in a dominant-negative fashion, leading to a variety of cellular and molecular changes, including nuclear abnormalities, defective DNA damage response (DDR) and DNA repair, and accelerated telomere attrition. Intriguingly, many of the manifestations of the HGPS cells are shared with normal aging cells. However, at a clinical level, HGPS does not fully match normal aging because of the accelerated nature of the phenotypes and its primary effects on connective tissues. Furthermore, the epigenetic changes in HGPS patients are of great interest and may play a crucial role in the pathogenesis of HGPS. Finally, various treatments for the HGPS patients have been developed in recent years with important effects at a cellular level, which translate to symptomatic improvement and increased lifespan.
Keywords: HGPS; Hutchinson–Gilford progeria syndrome; aging; laminopathy; progeria.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

