SCP4ssd: A Serverless Platform for Nucleotide Sequence Synthesis Difficulty Prediction Using an AutoML Model
- PMID: 36980878
- PMCID: PMC10048150
- DOI: 10.3390/genes14030605
SCP4ssd: A Serverless Platform for Nucleotide Sequence Synthesis Difficulty Prediction Using an AutoML Model
Abstract
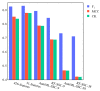
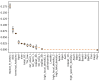
DNA synthesis is widely used in synthetic biology to construct and assemble sequences ranging from short RBS to ultra-long synthetic genomes. Many sequence features, such as the GC content and repeat sequences, are known to affect the synthesis difficulty and subsequently the synthesis cost. In addition, there are latent sequence features, especially local characteristics of the sequence, which might affect the DNA synthesis process as well. Reliable prediction of the synthesis difficulty for a given sequence is important for reducing the cost, but this remains a challenge. In this study, we propose a new automated machine learning (AutoML) approach to predict the DNA synthesis difficulty, which achieves an F1 score of 0.930 and outperforms the current state-of-the-art model. We found local sequence features that were neglected in previous methods, which might also affect the difficulty of DNA synthesis. Moreover, experimental validation based on ten genes of Escherichia coli strain MG1655 shows that our model can achieve an 80% accuracy, which is also better than the state of art. Moreover, we developed the cloud platform SCP4SSD using an entirely cloud-based serverless architecture for the convenience of the end users.
Keywords: AutoML; DNA synthesis; cloud platform; feature reduction; machine learning.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

