DNA Barcoding, Phylogenetic Analysis and Secondary Structure Predictions of Nepenthes ampullaria, Nepenthes gracilis and Nepenthes rafflesiana
- PMID: 36980969
- PMCID: PMC10048361
- DOI: 10.3390/genes14030697
DNA Barcoding, Phylogenetic Analysis and Secondary Structure Predictions of Nepenthes ampullaria, Nepenthes gracilis and Nepenthes rafflesiana
Abstract
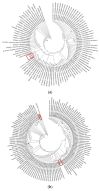
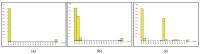
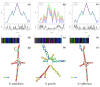
Nepentheceae, the most prominent carnivorous family in the Caryophyllales order, comprises the Nepenthes genus, which has modified leaf trap characteristics. Although most Nepenthes species have unique morphologies, their vegetative stages are identical, making identification based on morphology difficult. DNA barcoding is seen as a potential tool for plant identification, with small DNA segments amplified for species identification. In this study, three barcode loci; ribulose-bisphosphate carboxylase (rbcL), intergenic spacer 1 (ITS1) and intergenic spacer 2 (ITS2) and the usefulness of the ITS1 and ITS2 secondary structure for the molecular identification of Nepenthes species were investigated. An analysis of barcodes was conducted using BLASTn, pairwise genetic distance and diversity, followed by secondary structure prediction. The findings reveal that PCR and sequencing were both 100% successful. The present study showed the successful amplification of all targeted DNA barcodes at different sizes. Among the three barcodes, rbcL was the least efficient as a DNA barcode compared to ITS1 and ITS2. The ITS1 nucleotide analysis revealed that the ITS1 barcode had more variations compared to ITS2. The mean genetic distance (K2P) between them was higher for interspecies compared to intraspecies. The results showed that the DNA barcoding gap existed among Nepenthes species, and differences in the secondary structure distinguish the Nepenthes. The secondary structure generated in this study was found to successfully discriminate between the Nepenthes species, leading to enhanced resolutions.
Keywords: DNA barcode; ITS1; ITS2; Nepenthes; barcoding gap; phylogenetic; rbcL; secondary structure predictions.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- McPherson S., Robinson A.S. Field Guide to the Pitcher Plants of Peninsular Malaysia and Indochina. Redfern Natural History; Poole, UK: 2012.
-
- Clarke C. Nepenthes of Borneo. Natural History Publications (Borneo); Kota Kinabalu, Malaysia: 1997.
-
- Clarke C. Nepenthes of Sumatra and Peninsular Malaysia. Natural History Publications (Borneo); Kota Kinabalu, Malaysia: 2001. Nepenthes of Sumatra, and Peninsular Malaysia; p. 336.
-
- Cheek M., Jebb M. Flora Malesiana. National Herbarium of the Netherlands; Wageningen, The Netherlands: 2001. Flora Malesiana, Series 1: Volume 15: Nepenthaceae.
-
- Jebb M., Cheek M. A skeletal revision of nepenthes (Nepenthaceae) Blumea J. Plant Taxon. Plant Geogr. 1997;42:1–106.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

