Past Connectivity but Recent Inbreeding in Cross River Gorillas Determined Using Whole Genomes from Single Hairs
- PMID: 36981014
- PMCID: PMC10048488
- DOI: 10.3390/genes14030743
Past Connectivity but Recent Inbreeding in Cross River Gorillas Determined Using Whole Genomes from Single Hairs
Abstract
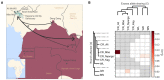
The critically endangered western gorillas (Gorilla gorilla) are divided into two subspecies: the western lowland (G. g. gorilla) and the Cross River (G. g. diehli) gorilla. Given the difficulty in sampling wild great ape populations and the small estimated size of the Cross River gorilla population, only one whole genome of a Cross River gorilla has been sequenced to date, hindering the study of this subspecies at the population level. In this study, we expand the number of whole genomes available for wild western gorillas, generating 41 new genomes (25 belonging to Cross River gorillas) using single shed hairs collected from gorilla nests. By combining these genomes with publicly available wild gorilla genomes, we confirm that Cross River gorillas form three population clusters. We also found little variation in genome-wide heterozygosity among them. Our analyses reveal long runs of homozygosity (>10 Mb), indicating recent inbreeding in Cross River gorillas. This is similar to that seen in mountain gorillas but with a much more recent bottleneck. We also detect past gene flow between two Cross River sites, Afi Mountain Wildlife Sanctuary and the Mbe Mountains. Furthermore, we observe past allele sharing between Cross River gorillas and the northern western lowland gorilla sites, as well as with the eastern gorilla species. This is the first study using single shed hairs from a wild species for whole genome sequencing to date. Taken together, our results highlight the importance of implementing conservation measures to increase connectivity among Cross River gorilla sites.
Keywords: Cross River gorilla; NGS; bottleneck; gene flow; hairs; inbreeding; non-invasive; wild gorillas.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Strindberg S., Maisels F., Williamson E.A., Blake S., Stokes E.J., Aba’a R., Abitsi G., Agbor A., Ambahe R.D., Bakabana P.C., et al. Guns, Germs, and Trees Determine Density and Distribution of Gorillas and Chimpanzees in Western Equatorial Africa. Sci. Adv. 2018;4:eaar2964. doi: 10.1126/sciadv.aar2964. - DOI - PMC - PubMed
-
- Dunn A., Bergl R., Byler D., Eben-Ebai S., Ndeloh Etiendem D., Fotso R., Ikfuingei R., Imong I., Jameson C., Macfie L., et al. Revised Regional Action Plan for the Conservation of the Cross River Gorilla (Gorilla gorilla diehli) 2014–2019. IUCN/SSC Primate Specialist Group and Wildlife Conservation Society; New York, NY, USA: 2014.
-
- Bergl R.A., Warren Y., Nicholas A., Dunn A., Imong I., Sunderland-Groves J.L., Oates J.F. Remote Sensing Analysis Reveals Habitat, Dispersal Corridors and Expanded Distribution for the Critically Endangered Cross River Gorilla Gorilla gorilla diehli. Oryx. 2012;46:278–289. doi: 10.1017/S0030605310001857. - DOI
-
- Oates J., Sunderland-Groves J., Bergl R., Dunn A., Nicholas A., Takang E., Omeni F., Imong I., Fotso R., Nkembi L., et al. Regional Action Plan for the Conservation of the Cross River Gorilla (Gorilla gorilla diehli) IUCN/SSC Primate Specialist Group and Wildlife Conservation Society; Arlington, VA, USA: 2007.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

