Coupling Hydrophilic Interaction Chromatography and Reverse-Phase Chromatography for Improved Direct Analysis of Grape Seed Proanthocyanidins
- PMID: 36981246
- PMCID: PMC10048310
- DOI: 10.3390/foods12061319
Coupling Hydrophilic Interaction Chromatography and Reverse-Phase Chromatography for Improved Direct Analysis of Grape Seed Proanthocyanidins
Abstract
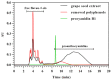
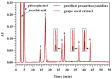
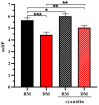
Acid-catalyzed depolymerization is recognized as the most practical method for analyzing subunit composition and the polymerization degree of proanthocyanidins, involving purification by removing free flavan-3-ols, as well as acid-catalyzed cleavage and the identification of cleavage products. However, after the removal of proanthocyanidins with low molecular weights during purification, the formation of anthocyanidins from the extension subunits accompanying acid-catalyzed cleavage occurred. Thus, grape seed extract other than purified proanthocyanidins was applied to acid-catalyzed depolymerization. Hydrophilic interaction chromatography was developed to quantify free flavan-3-ols in grape seed extract to distinguish them from flavan-3-ols from terminal subunits of proanthocyanidins. Reverse-phase chromatography was used to analyze anthocyanidins and cleavage products at 550 and 280 nm, respectively. It is found that the defects of the recognized method did not influence the results of the subunit composition, but both altered the mean degree of polymerization. The established method was able to directly analyze proanthocyanidins in grape seed extract for higher accuracy and speed than the recognized method.
Keywords: anthocyanidin; direct analysis; hydrophilic interaction chromatography; mean degree of polymerization; proanthocyanidins.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Gu L., Kelm M.A., Hammerstone J.F., Beecher G., Holden J., Haytowitz D., Gebhardt S., Prior R.L. Concentrations of proanthocyanidins in common foods and estimations of normal consumption. J. Nutr. 2004;134:613–617. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

