Data-Independent Acquisition Proteomics Reveals the Effects of Red and Blue Light on the Growth and Development of Moso Bamboo (Phyllostachys edulis) Seedlings
- PMID: 36982175
- PMCID: PMC10049362
- DOI: 10.3390/ijms24065103
Data-Independent Acquisition Proteomics Reveals the Effects of Red and Blue Light on the Growth and Development of Moso Bamboo (Phyllostachys edulis) Seedlings
Abstract
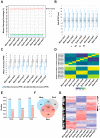
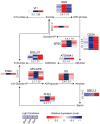
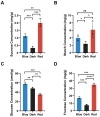
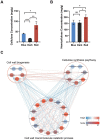
Moso bamboo is a rapidly growing species with significant economic, social, and cultural value. Transplanting moso bamboo container seedlings for afforestation has become a cost-effective method. The growth and development of the seedlings is greatly affected by the quality of light, including light morphogenesis, photosynthesis, and secondary metabolite production. Therefore, studies on the effects of specific light wavelengths on the physiology and proteome of moso bamboo seedlings are crucial. In this study, moso bamboo seedlings were germinated in darkness and then exposed to blue and red light conditions for 14 days. The effects of these light treatments on seedling growth and development were observed and compared through proteomics analysis. Results showed that moso bamboo has higher chlorophyll content and photosynthetic efficiency under blue light, while it displays longer internode and root length, more dry weight, and higher cellulose content under red light. Proteomics analysis reveals that these changes under red light are likely caused by the increased content of cellulase CSEA, specifically expressed cell wall synthetic proteins, and up-regulated auxin transporter ABCB19 in red light. Additionally, blue light is found to promote the expression of proteins constituting photosystem II, such as PsbP and PsbQ, more than red light. These findings provide new insights into the growth and development of moso bamboo seedlings regulated by different light qualities.
Keywords: blue light; data-independent acquisition proteomics; growth and development; moso bamboo; red light.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Ramakrishnan M., Yrjälä K., Vinod K.K., Sharma A., Cho J., Satheesh V., Zhou M. Genetics and genomics of moso bamboo (phyllostachys edulis): Current status, future challenges, and biotechnological opportunities toward a sustainable bamboo industry. Food Energy Secur. 2020;9:e229. doi: 10.1002/fes3.229. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

