Genetic Networks of Alzheimer's Disease, Aging, and Longevity in Humans
- PMID: 36982253
- PMCID: PMC10049434
- DOI: 10.3390/ijms24065178
Genetic Networks of Alzheimer's Disease, Aging, and Longevity in Humans
Abstract
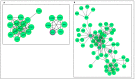
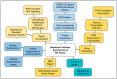
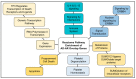
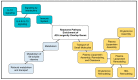
Human genomic analysis and genome-wide association studies (GWAS) have identified genes that are risk factors for early and late-onset Alzheimer's disease (AD genes). Although the genetics of aging and longevity have been extensively studied, previous studies have focused on a specific set of genes that have been shown to contribute to or are a risk factor for AD. Thus, the connections among the genes involved in AD, aging, and longevity are not well understood. Here, we identified the genetic interaction networks (referred to as pathways) of aging and longevity within the context of AD by using a gene set enrichment analysis by Reactome that cross-references more than 100 bioinformatic databases to allow interpretation of the biological functions of gene sets through a wide variety of gene networks. We validated the pathways with a threshold of p-value < 1.00 × 10-5 using the databases to extract lists of 356 AD genes, 307 aging-related (AR) genes, and 357 longevity genes. There was a broad range of biological pathways involved in AR and longevity genes shared with AD genes. AR genes identified 261 pathways within the threshold of p < 1.00 × 10-5, of which 26 pathways (10% of AR gene pathways) were further identified by overlapping genes among AD and AR genes. The overlapped pathways included gene expression (p = 4.05 × 10-11) including ApoE, SOD2, TP53, and TGFB1 (p = 2.84 × 10-10); protein metabolism and SUMOylation, including E3 ligases and target proteins (p = 1.08 × 10-7); ERBB4 signal transduction (p = 2.69 × 10-6); the immune system, including IL-3 and IL-13 (p = 3.83 × 10-6); programmed cell death (p = 4.36 × 10-6); and platelet degranulation (p = 8.16 × 10-6), among others. Longevity genes identified 49 pathways within the threshold, of which 12 pathways (24% of longevity gene pathways) were further identified by overlapping genes among AD and longevity genes. They include the immune system, including IL-3 and IL-13 (p = 7.64 × 10-8), plasma lipoprotein assembly, remodeling and clearance (p < 4.02 × 10-6), and the metabolism of fat-soluble vitamins (p = 1.96 × 10-5). Thus, this study provides shared genetic hallmarks of aging, longevity, and AD backed up by statistical significance. We discuss the significant genes involved in these pathways, including TP53, FOXO, SUMOylation, IL4, IL6, APOE, and CEPT, and suggest that mapping the gene network pathways provide a useful basis for further medical research on AD and healthy aging.
Keywords: age-related comorbidity; centenarian; dementia; epigenetics; hallmark of aging; life extension; longevity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Matthews K.A., Xu W., Gaglioti A.H., Holt J.B., Croft J.B., Mack D., McGuire L.C. Racial and ethnic estimates of Alzheimer’s disease and related dementias in the United States (2015–2060) in adults aged ≥65 years. Alzheimer’s Dement. 2019;15:17–24. doi: 10.1016/j.jalz.2018.06.3063. - DOI - PMC - PubMed
-
- Goate A., Chartier-Harlin M.-C., Mullan M., Brown J., Crawford F., Fidani L., Giuffra L., Haynes A., Irving N., James L., et al. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer’s disease. Nature. 1991;349:704–706. doi: 10.1038/349704a0. - DOI - PubMed
-
- Rogaev E.I., Sherrington R., Rogaeva E.A., Levesque G., Ikeda M., Liang Y., Chi H., Lin C., Holman K., Tsuda T., et al. Familial Alzheimer’s disease in kindreds with missense mutations in a gene on chromosome 1 related to the Alzheimer’s disease type 3 gene. Nature. 1995;376:775–778. doi: 10.1038/376775a0. - DOI - PubMed
-
- Sherva S., Kowall N.W. Genetics of Alzheimer Disease. 2022. [(accessed on 6 January 2023)]. Available online: https://www-uptodate-com.eu1.proxy.openathens.net.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

