Biology and Regulation of Staphylococcal Biofilm
- PMID: 36982293
- PMCID: PMC10049468
- DOI: 10.3390/ijms24065218
Biology and Regulation of Staphylococcal Biofilm
Abstract
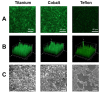
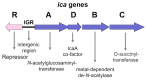
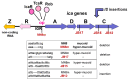
Despite continuing progress in medical and surgical procedures, staphylococci remain the major Gram-positive bacterial pathogens that cause a wide spectrum of diseases, especially in patients requiring the utilization of indwelling catheters and prosthetic devices implanted temporarily or for prolonged periods of time. Within the genus, if Staphylococcus aureus and S. epidermidis are prevalent species responsible for infections, several coagulase-negative species which are normal components of our microflora also constitute opportunistic pathogens that are able to infect patients. In such a clinical context, staphylococci producing biofilms show an increased resistance to antimicrobials and host immune defenses. Although the biochemical composition of the biofilm matrix has been extensively studied, the regulation of biofilm formation and the factors contributing to its stability and release are currently still being discovered. This review presents and discusses the composition and some regulation elements of biofilm development and describes its clinical importance. Finally, we summarize the numerous and various recent studies that address attempts to destroy an already-formed biofilm within the clinical context as a potential therapeutic strategy to avoid the removal of infected implant material, a critical event for patient convenience and health care costs.
Keywords: biofilm; foreign body; gene expression; metabolism; phenotype; regulation; resistance; staphylococci; tolerance.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

