Epigenetic Regulation Mediated by Sphingolipids in Cancer
- PMID: 36982369
- PMCID: PMC10048860
- DOI: 10.3390/ijms24065294
Epigenetic Regulation Mediated by Sphingolipids in Cancer
Abstract
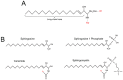
Epigenetic changes are heritable modifications that do not directly affect the DNA sequence. In cancer cells, the maintenance of a stable epigenetic profile can be crucial to support survival and proliferation, and said profile can differ significantly from that of healthy cells. The epigenetic profile of a cancer cell can be modulated by several factors, including metabolites. Recently, sphingolipids have emerged as novel modulators of epigenetic changes. Ceramide and sphingosine 1-phosphate have become well known in cancer due to activating anti-tumour and pro-tumour signalling pathways, respectively, and they have recently been shown to also induce several epigenetic modifications connected to cancer growth. Additionally, acellular factors in the tumour microenvironment, such as hypoxia and acidosis, are now recognised as crucial in promoting aggressiveness through several mechanisms, including epigenetic modifications. Here, we review the existing literature on sphingolipids, cancer, and epigenetic changes, with a focus on the interaction between these elements and components of the chemical tumour microenvironment.
Keywords: acidosis; bone cancer; cancer; epigenetics; hypoxia; sphingolipids; tumour microenvironment.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

