Transcriptomic Signatures of Single-Suture Craniosynostosis Phenotypes
- PMID: 36982425
- PMCID: PMC10049207
- DOI: 10.3390/ijms24065353
Transcriptomic Signatures of Single-Suture Craniosynostosis Phenotypes
Abstract
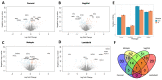
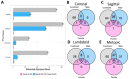
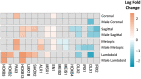
Craniosynostosis is a birth defect where calvarial sutures close prematurely, as part of a genetic syndrome or independently, with unknown cause. This study aimed to identify differences in gene expression in primary calvarial cell lines derived from patients with four phenotypes of single-suture craniosynostosis, compared to controls. Calvarial bone samples (N = 388 cases/85 controls) were collected from clinical sites during reconstructive skull surgery. Primary cell lines were then derived from the tissue and used for RNA sequencing. Linear models were fit to estimate covariate adjusted associations between gene expression and four phenotypes of single-suture craniosynostosis (lambdoid, metopic, sagittal, and coronal), compared to controls. Sex-stratified analysis was also performed for each phenotype. Differentially expressed genes (DEGs) included 72 genes associated with coronal, 90 genes associated with sagittal, 103 genes associated with metopic, and 33 genes associated with lambdoid craniosynostosis. The sex-stratified analysis revealed more DEGs in males (98) than females (4). There were 16 DEGs that were homeobox (HOX) genes. Three TFs (SUZ12, EZH2, AR) significantly regulated expression of DEGs in one or more phenotypes. Pathway analysis identified four KEGG pathways associated with at least one phenotype of craniosynostosis. Together, this work suggests unique molecular mechanisms related to craniosynostosis phenotype and fetal sex.
Keywords: RNA sequencing; craniosynostosis; homeobox; transcriptome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Wilkie A.O.M., Byren J.C., Hurst J.A., Jayamohan J., Johnson D., Knight S.J.L., Lester T., Richards P.G., Twigg S.R.F., Wall S.A. Prevalence and Complications of Single-Gene and Chromosomal Disorders in Craniosynostosis. Pediatrics. 2010;126:e391–e400. doi: 10.1542/peds.2009-3491. - DOI - PMC - PubMed
-
- Kabbani H., Raghuveer T.S. Craniosynostosis. Am. Fam. Physician. 2004;69:2863–2870. - PubMed
-
- Ye X., Guilmatre A., Reva B., Peter I., Heuzé Y., Richtsmeier J.T., Fox D.J., Goedken R.J., Jabs E.W., Romitti P.A. Mutation Screening of Candidate Genes in Patients with Nonsyndromic Sagittal Craniosynostosis. Plast. Reconstr. Surg. 2016;137:952–961. doi: 10.1097/01.prs.0000479978.75545.ee. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

