Transcriptomic Differentiation of Phenotypes in Chronic Rhinosinusitis and Its Implications for Understanding the Underlying Mechanisms
- PMID: 36982612
- PMCID: PMC10051401
- DOI: 10.3390/ijms24065541
Transcriptomic Differentiation of Phenotypes in Chronic Rhinosinusitis and Its Implications for Understanding the Underlying Mechanisms
Abstract
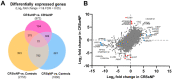
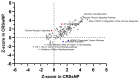
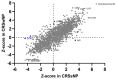
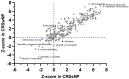
Chronic rhinosinusitis (CRS) is a multifaceted disease with variable clinical courses and outcomes. We aimed to determine CRS-associated nasal-tissue transcriptome in clinically well-characterized and phenotyped individuals, to gain a novel insight into the biological pathways of the disease. RNA-sequencing of tissue samples of patients with CRS with polyps (CRSwNP), without polyps (CRSsNP), and controls were performed. Characterization of differently expressed genes (DEGs) and functional and pathway analysis was undertaken. We identified 782 common CRS-associated nasal-tissue DEGs, while 375 and 328 DEGs were CRSwNP- and CRSsNP-specific, respectively. Common key DEGs were found to be involved in dendritic cell maturation, the neuroinflammation pathway, and the inhibition of the matrix metalloproteinases. Distinct CRSwNP-specific DEGs were involved in NF-kβ canonical pathways, Toll-like receptor signaling, HIF1α regulation, and the Th2 pathway. CRSsNP involved the NFAT pathway and changes in the calcium pathway. Our findings offer new insights into the common and distinct molecular mechanisms underlying CRSwNP and CRSsNP, providing further understanding of the complex pathophysiology of the CRS, with future research directions for novel treatment strategies.
Keywords: RNA; phenotype; sequence analysis; sinusitis; transcriptome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Tc17/IL-17A Up-Regulated the Expression of MMP-9 via NF-κB Pathway in Nasal Epithelial Cells of Patients With Chronic Rhinosinusitis.Front Immunol. 2018 Sep 19;9:2121. doi: 10.3389/fimmu.2018.02121. eCollection 2018. Front Immunol. 2018. PMID: 30283454 Free PMC article.
-
Differing roles for TGF-β/Smad signaling in osteitis in chronic rhinosinusitis with and without nasal polyps.Am J Rhinol Allergy. 2015 Sep-Oct;29(5):e152-9. doi: 10.2500/ajra.2015.29.4241. Epub 2015 Aug 10. Am J Rhinol Allergy. 2015. PMID: 26265084
-
Tissue Eosinophilia is Superior to an Analysis by Polyp Status for the Chronic Rhinosinusitis Transcriptome: An RNA Study.Laryngoscope. 2023 Oct;133(10):2480-2489. doi: 10.1002/lary.30544. Epub 2023 Jan 3. Laryngoscope. 2023. PMID: 36594502
-
Differential expression and role of S100 proteins in chronic rhinosinusitis.Curr Opin Allergy Clin Immunol. 2020 Feb;20(1):14-22. doi: 10.1097/ACI.0000000000000595. Curr Opin Allergy Clin Immunol. 2020. PMID: 31644435 Free PMC article. Review.
-
Phenotypes in Chronic Rhinosinusitis.Curr Allergy Asthma Rep. 2020 May 19;20(7):20. doi: 10.1007/s11882-020-00916-6. Curr Allergy Asthma Rep. 2020. PMID: 32430653 Review.
Cited by
-
Host-microbe interactions in chronic rhinosinusitis biofilms and models for investigation.Biofilm. 2023 Sep 29;6:100160. doi: 10.1016/j.bioflm.2023.100160. eCollection 2023 Dec 15. Biofilm. 2023. PMID: 37928619 Free PMC article. Review.
-
Trends and future directions in chronic rhinosinusitis with nasal polyps: A bibliometric analysis.Braz J Otorhinolaryngol. 2025 Jul 3;91(5):101672. doi: 10.1016/j.bjorl.2025.101672. Online ahead of print. Braz J Otorhinolaryngol. 2025. PMID: 40614574 Free PMC article.
References
-
- Wang X., Zhang N., Bo M., Holtappels G., Zheng M., Lou H., Wang H., Zhang L., Bachert C. Diversity of TH Cytokine Profiles in Patients with Chronic Rhinosinusitis: A Multicenter Study in Europe, Asia, and Oceania. J. Allergy Clin. Immunol. 2016;138:1344–1353. doi: 10.1016/j.jaci.2016.05.041. - DOI - PubMed
-
- Tomassen P., Vandeplas G., Van Zele T., Cardell L.O., Arebro J., Olze H., Förster-Ruhrmann U., Kowalski M.L., Olszewska-Ziaber A., Holtappels G., et al. Inflammatory Endotypes of Chronic Rhinosinusitis Based on Cluster Analysis of Biomarkers. J. Allergy Clin. Immunol. 2016;137:1449–1456e4. doi: 10.1016/j.jaci.2015.12.1324. - DOI - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

