Diapause-Linked Gene Expression Pattern and Related Candidate Duplicated Genes of the Mountain Butterfly Parnassius glacialis (Lepidoptera: Papilionidae) Revealed by Comprehensive Transcriptome Profiling
- PMID: 36982649
- PMCID: PMC10058462
- DOI: 10.3390/ijms24065577
Diapause-Linked Gene Expression Pattern and Related Candidate Duplicated Genes of the Mountain Butterfly Parnassius glacialis (Lepidoptera: Papilionidae) Revealed by Comprehensive Transcriptome Profiling
Abstract
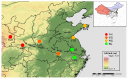
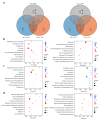
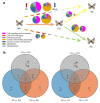
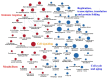
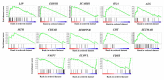
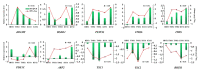
The mountain butterfly Parnassius glacialis is a representative species of the genus Parnassius, which probably originated in the high-altitude Qinhai-Tibet Plateau in the Miocene and later dispersed eastward into relatively low-altitude regions of central to eastern China. However, little is known about the molecular mechanisms underlying the long-term evolutionary adaptation to heterogeneous environmental conditions of this butterfly species. In this study, we obtained the high-throughput RNA-Seq data from twenty-four adult individuals in eight localities, covering nearly all known distributional areas in China, and firstly identified the diapause-linked gene expression pattern that is likely to correlate with local adaptation in adult P. glacialis populations. Secondly, we found a series of pathways responsible for hormone biosynthesis, energy metabolism and immune defense that also exhibited unique enrichment patterns in each group that are probably related to habitat-specific adaptability. Furthermore, we also identified a suite of duplicated genes (including two transposable elements) that are mostly co-expressed to promote the plastic responses to different environmental conditions. Together, these findings can help us to better understand this species' successful colonization to distinct geographic areas from the western to eastern areas of China, and also provide us with some insights into the evolution of diapause in mountain Parnassius butterfly species.
Keywords: Parnassius glacialis; Transcriptome; diapause; environmental adaptation; genome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

