DNA Methylation of Window of Implantation Genes in Cervical Secretions Predicts Ongoing Pregnancy in Infertility Treatment
- PMID: 36982674
- PMCID: PMC10051225
- DOI: 10.3390/ijms24065598
DNA Methylation of Window of Implantation Genes in Cervical Secretions Predicts Ongoing Pregnancy in Infertility Treatment
Abstract
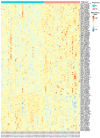
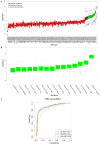
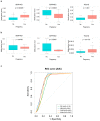
Window of implantation (WOI) genes have been comprehensively identified at the single cell level. DNA methylation changes in cervical secretions are associated with in vitro fertilization embryo transfer (IVF-ET) outcomes. Using a machine learning (ML) approach, we aimed to determine which methylation changes in WOI genes from cervical secretions best predict ongoing pregnancy during embryo transfer. A total of 2708 promoter probes were extracted from mid-secretory phase cervical secretion methylomic profiles for 158 WOI genes, and 152 differentially methylated probes (DMPs) were selected. Fifteen DMPs in 14 genes (BMP2, CTSA, DEFB1, GRN, MTF1, SERPINE1, SERPINE2, SFRP1, STAT3, TAGLN2, TCF4, THBS1, ZBTB20, ZNF292) were identified as the most relevant to ongoing pregnancy status. These 15 DMPs yielded accuracy rates of 83.53%, 85.26%, 85.78%, and 76.44%, and areas under the receiver operating characteristic curves (AUCs) of 0.90, 0.91, 0.89, and 0.86 for prediction by random forest (RF), naïve Bayes (NB), support vector machine (SVM), and k-nearest neighbors (KNN), respectively. SERPINE1, SERPINE2, and TAGLN2 maintained their methylation difference trends in an independent set of cervical secretion samples, resulting in accuracy rates of 71.46%, 80.06%, 80.72%, and 80.68%, and AUCs of 0.79, 0.84, 0.83, and 0.82 for prediction by RF, NB, SVM, and KNN, respectively. Our findings demonstrate that methylation changes in WOI genes detected noninvasively from cervical secretions are potential markers for predicting IVF-ET outcomes. Further studies of cervical secretion of DNA methylation markers may provide a novel approach for precision embryo transfer.
Keywords: DNA methylation; boruta algorithm; cervical secretion; endometrial receptivity; in vitro fertilization; machine learning; noninvasive; window of implantation.
Conflict of interest statement
The application of these DNA methylation biomarkers is patent pending.
Figures
References
-
- De Geyter C., Wyns C., Calhaz-Jorge C., De Mouzon J., Ferraretti A.P., Kupka M., Andersen A.N., Nygren K.G., Goossens V. 20 years of the European IVF-monitoring Consortium registry: What have we learned? A comparison with registries from two other regions. Hum. Reprod. 2020;35:2832–2849. doi: 10.1093/humrep/deaa250. - DOI - PMC - PubMed
-
- Munné S., Kaplan B., Frattarelli J.L., Child T., Nakhuda G., Shamma F.N., Silverberg K., Kalista T., Handyside A.H., Katz-Jaffe M., et al. Preimplantation genetic testing for aneuploidy versus morphology as selection criteria for single fro-zen-thawed embryo transfer in good-prognosis patients: A multicenter randomized clinical trial. Fertil. Steril. 2019;112:1071–1079.e7. doi: 10.1016/j.fertnstert.2019.07.1346. - DOI - PubMed
-
- Navot D., Bergh P.A., Williams M., Garrlsi G.J., Guzman I., Sandler B., Fox J., Schreiner-Engel P., Hofmann G.E., Grunfeld L. An Insight into Early Reproductive Processes through the In Vivo Model of Ovum Donation. J. Clin. Endocrinol. Metab. 1991;72:408–414. doi: 10.1210/jcem-72-2-408. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

