Shared Genes of PPARG and NOS2 in Alzheimer's Disease and Ulcerative Colitis Drive Macrophages and Microglia Polarization: Evidence from Bioinformatics Analysis and Following Validation
- PMID: 36982725
- PMCID: PMC10058634
- DOI: 10.3390/ijms24065651
Shared Genes of PPARG and NOS2 in Alzheimer's Disease and Ulcerative Colitis Drive Macrophages and Microglia Polarization: Evidence from Bioinformatics Analysis and Following Validation
Abstract
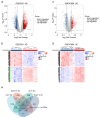
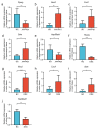
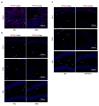
Emerging evidence shows that peripheral systemic inflammation, such as inflammatory bowel disease (IBD), has a close even interaction with central nervous disorders such as Alzheimer's disease (AD). This study is designed to further clarify the relationship between AD and ulcerative colitis (UC, a subclass of IBD). The GEO database was used to download gene expression profiles for AD (GSE5281) and UC (GSE47908). Bioinformatics analysis included GSEA, KEGG pathway, Gene Ontology (GO), WikiPathways, PPI network, and hub gene identification. After screening the shared genes, qRT-PCR, Western blot, and immunofluorescence were used to verify the reliability of the dataset and further confirm the shared genes. GSEA, KEGG, GO, and WikiPathways suggested that PPARG and NOS2 were identified as shared genes and hub genes by cytoHubba in AD and UC and further validated via qRT-PCR and Western blot. Our work identified PPARG and NOS2 are shared genes of AD and UC. They drive macrophages and microglia heterogeneous polarization, which may be potential targets for treating neural dysfunction induced by systemic inflammation and vice versa.
Keywords: Alzheimer’s disease; NOS2; PPARG; bioinformatics; shared gene; ulcerative colitis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures








References
-
- Marizzoni M., Cattaneo A., Mirabelli P., Festari C., Lopizzo N., Nicolosi V., Mombelli E., Mazzelli M., Luongo D., Naviglio D., et al. Short-Chain Fatty Acids and Lipopolysaccharide as Mediators Between Gut Dysbiosis and Amyloid Pathology in Alzheimer’s Disease. J. Alzheimer’s Dis. 2020;78:683–697. doi: 10.3233/JAD-200306. - DOI - PubMed
-
- Liao P.H., Chiang H.L., Shun C.T., Hang J.F., Chiu H.M., Wu M.S., Lin C.H. Colonic Leucine-Rich Repeat Kinase 2 Expression Is Increased and Associated With Disease Severity in Patients With Parkinson’s Disease. Front. Aging Neurosci. 2021;13:819373. doi: 10.3389/fnagi.2021.819373. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- 2022YFC3500703/the National Key R&D Program of China
- 2019YFC1709001/the National Key R&D Program of China
- 82174512/the National Natural Science Foundation of China
- 81873383/the National Natural Science Foundation of China
- ZYYCXTD-D-202003/the Innovation Team and Talents Cultivation Program of National Administration of Traditional Chinese Medicine
LinkOut - more resources
Full Text Sources
Medical

