Kinetic Characterization and Catalytic Mechanism of N-Acetylornithine Aminotransferase Encoded by slr1022 Gene from Synechocystis sp. PCC6803
- PMID: 36982927
- PMCID: PMC10057298
- DOI: 10.3390/ijms24065853
Kinetic Characterization and Catalytic Mechanism of N-Acetylornithine Aminotransferase Encoded by slr1022 Gene from Synechocystis sp. PCC6803
Abstract
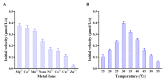
The enzyme encoded by slr1022 gene from Synechocystis sp. PCC6803 was reported to function as N-acetylornithine aminotransferase, γ-aminobutyric acid aminotransferase, and ornithine aminotransferase, which played important roles in multiple metabolic pathways. Among these functions, N-acetylornithine aminotransferase catalyzes the reversible conversion of N-acetylornithine to N-acetylglutamate-5-semialdehyde with PLP as cofactor, which is a key step in the arginine biosynthesis pathway. However, the investigation of the detailed kinetic characteristics and catalytic mechanism of Slr1022 has not been carried out yet. In this study, the exploration of kinetics of recombinant Slr1022 illustrated that Slr1022 mainly functioned as N-acetylornithine aminotransferase with low substrate specificity to γ-aminobutyric acid and ornithine. Kinetic assay of Slr1022 variants and the model structure of Slr1022 with N-acetylornithine-PLP complex revealed that Lys280 and Asp251 residues were the key amino acids of Slr1022. The respective mutation of the above two residues to Ala resulted in the activity depletion of Slr1022. Meanwhile, Glu223 residue was involved in substrate binding and it served as a switch between the two half reactions. Other residues such as Thr308, Gln254, Tyr39, Arg163, and Arg402 implicated a substrate recognition and catalytic process of the reaction. The results of this study further enriched the understanding of the catalytic kinetics and mechanism of N-acetylornithine aminotransferase, especially from cyanobacteria.
Keywords: N-acetylornithine aminotransferase; Synechocystis sp. PCC6803; catalytic mechanism; kinetic characterization; site-directed mutagenesis.
Conflict of interest statement
The authors declare no conflict of interests.
Figures





References
-
- Heyl D., Luz E., Harris S.A., Folker K. Phosphates of the Vitamin B, Group. I. The Structure of Codecarboxylase. J. Am. Chem. Soc. 1951;73:3430–3433. doi: 10.1021/ja01151a126. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

