Co-Expression Network Analysis Identifies Molecular Determinants of Loneliness Associated with Neuropsychiatric and Neurodegenerative Diseases
- PMID: 36982982
- PMCID: PMC10058494
- DOI: 10.3390/ijms24065909
Co-Expression Network Analysis Identifies Molecular Determinants of Loneliness Associated with Neuropsychiatric and Neurodegenerative Diseases
Abstract
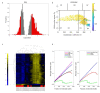
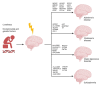
Loneliness and social isolation are detrimental to mental health and may lead to cognitive impairment and neurodegeneration. Although several molecular signatures of loneliness have been identified, the molecular mechanisms by which loneliness impacts the brain remain elusive. Here, we performed a bioinformatics approach to untangle the molecular underpinnings associated with loneliness. Co-expression network analysis identified molecular 'switches' responsible for dramatic transcriptional changes in the nucleus accumbens of individuals with known loneliness. Loneliness-related switch genes were enriched in cell cycle, cancer, TGF-β, FOXO, and PI3K-AKT signaling pathways. Analysis stratified by sex identified switch genes in males with chronic loneliness. Male-specific switch genes were enriched in infection, innate immunity, and cancer-related pathways. Correlation analysis revealed that loneliness-related switch genes significantly overlapped with 82% and 68% of human studies on Alzheimer's (AD) and Parkinson's diseases (PD), respectively, in gene expression databases. Loneliness-related switch genes, BCAM, NECTIN2, NPAS3, RBM38, PELI1, DPP10, and ASGR2, have been identified as genetic risk factors for AD. Likewise, switch genes HLA-DRB5, ALDOA, and GPNMB are known genetic loci in PD. Similarly, loneliness-related switch genes overlapped in 70% and 64% of human studies on major depressive disorder and schizophrenia, respectively. Nine switch genes, HLA-DRB5, ARHGAP15, COL4A1, RBM38, DMD, LGALS3BP, WSCD2, CYTH4, and CNTRL, overlapped with known genetic variants in depression. Seven switch genes, NPAS3, ARHGAP15, LGALS3BP, DPP10, SMYD3, CPXCR1, and HLA-DRB5 were associated with known risk factors for schizophrenia. Collectively, we identified molecular determinants of loneliness and dysregulated pathways in the brain of non-demented adults. The association of switch genes with known risk factors for neuropsychiatric and neurodegenerative diseases provides a molecular explanation for the observed prevalence of these diseases among lonely individuals.
Keywords: Alzheimer’s disease; Parkinson’s disease; loneliness; major depressive disorder; network analysis; neurodegenerative disease; neuropsychiatric diseases; schizophrenia; social isolation.
Conflict of interest statement
J.A.S is the founder of NeuroHub Analytics, LLC. J.P.Q. is the founder of Q Regulating Systems, LLC. The remaining author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Canli T., Yu L., Yu X., Zhao H., Fleischman D., Wilson R.S., De Jager P.L., Bennett D.A. Loneliness 5 years ante-mortem is associated with disease-related differential gene expression in postmortem dorsolateral prefrontal cortex. Transl. Psychiatry. 2018;8:2. doi: 10.1038/s41398-017-0086-2. - DOI - PMC - PubMed
-
- Beutel M.E., Klein E.M., Brähler E., Reiner I., Jünger C., Michal M., Wiltink J., Wild P.S., Münzel T., Lackner K.J., et al. Loneliness in the general population: Prevalence, determinants and relations to mental health. BMC Psychiatry. 2017;17:97. doi: 10.1186/s12888-017-1262-x. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

